Host-Specific and Segment-Specific Evolutionary Dynamics of Avian and Human Influenza A Viruses: A Systematic Review
- PMID: 26760775
- PMCID: PMC4720117
- DOI: 10.1371/journal.pone.0147021
Host-Specific and Segment-Specific Evolutionary Dynamics of Avian and Human Influenza A Viruses: A Systematic Review
Abstract
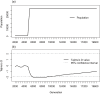
Understanding the evolutionary dynamics of influenza viruses is essential to control both avian and human influenza. Here, we analyze host-specific and segment-specific Tajima's D trends of influenza A virus through a systematic review using viral sequences registered in the National Center for Biotechnology Information. To avoid bias from viral population subdivision, viral sequences were stratified according to their sampling locations and sampling years. As a result, we obtained a total of 580 datasets each of which consists of nucleotide sequences of influenza A viruses isolated from a single population of hosts at a single sampling site within a single year. By analyzing nucleotide sequences in the datasets, we found that Tajima's D values of viral sequences were different depending on hosts and gene segments. Tajima's D values of viruses isolated from chicken and human samples showed negative, suggesting purifying selection or a rapid population growth of the viruses. The negative Tajima's D values in rapidly growing viral population were also observed in computer simulations. Tajima's D values of PB2, PB1, PA, NP, and M genes of the viruses circulating in wild mallards were close to zero, suggesting that these genes have undergone neutral selection in constant-sized population. On the other hand, Tajima's D values of HA and NA genes of these viruses were positive, indicating HA and NA have undergone balancing selection in wild mallards. Taken together, these results indicated the existence of unknown factors that maintain viral subtypes in wild mallards.
Conflict of interest statement
Figures
Similar articles
-
Genetic and Molecular Characterization of Avian Influenza A(H9N2) Viruses from Live Bird Markets (LBM) in Senegal.Viruses. 2025 Jan 8;17(1):73. doi: 10.3390/v17010073. Viruses. 2025. PMID: 39861862 Free PMC article.
-
Assessment of the public health risk of novel reassortant H3N3 avian influenza viruses that emerged in chickens.mBio. 2025 Jul 9;16(7):e0067725. doi: 10.1128/mbio.00677-25. Epub 2025 Jun 16. mBio. 2025. PMID: 40521889 Free PMC article.
-
Host origin is a determinant of coevolution between gene segments of avian H9 influenza viruses.J Virol. 2025 Jul 22;99(7):e0151824. doi: 10.1128/jvi.01518-24. Epub 2025 Jun 13. J Virol. 2025. PMID: 40511915 Free PMC article.
-
Avian influenza in Latin America: A systematic review of serological and molecular studies from 2000-2015.PLoS One. 2017 Jun 20;12(6):e0179573. doi: 10.1371/journal.pone.0179573. eCollection 2017. PLoS One. 2017. PMID: 28632771 Free PMC article.
-
A systematic review of mechanistic models used to study avian influenza virus transmission and control.Vet Res. 2023 Oct 18;54(1):96. doi: 10.1186/s13567-023-01219-0. Vet Res. 2023. PMID: 37853425 Free PMC article.
Cited by
-
Molecular Characterization and Analysis of Human Trichostrongylus Species in an Endemic Region of Iran Based on COX 1 Gene; A Cross-Sectional Study.Health Sci Rep. 2025 Apr 18;8(4):e70612. doi: 10.1002/hsr2.70612. eCollection 2025 Apr. Health Sci Rep. 2025. PMID: 40256140 Free PMC article.
-
Detection of homologous recombination events in SARS-CoV-2.Biotechnol Lett. 2022 Mar;44(3):399-414. doi: 10.1007/s10529-021-03218-7. Epub 2022 Jan 17. Biotechnol Lett. 2022. PMID: 35037234 Free PMC article.
-
Epidemiological, clinical, and virologic features of two family clusters of avian influenza A (H7N9) virus infections in Southeast China.Sci Rep. 2017 May 4;7(1):1512. doi: 10.1038/s41598-017-01761-w. Sci Rep. 2017. PMID: 28473725 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

