Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway
- PMID: 26761346
- PMCID: PMC4835958
- DOI: 10.1080/15548627.2015.1134081
Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway
Abstract
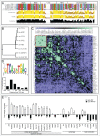
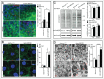
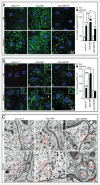
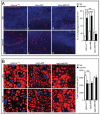
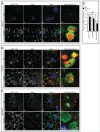
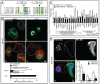
An evolutionarily conserved gene network regulates the expression of genes involved in lysosome biogenesis, autophagy, and lipid metabolism. In mammals, TFEB and other members of the MiTF-TFE family of transcription factors control this network. Here we report that the lysosomal-autophagy pathway is controlled by Mitf gene in Drosophila melanogaster. Mitf is the single MiTF-TFE family member in Drosophila and prior to this work was known only for its function in eye development. We show that Mitf regulates the expression of genes encoding V-ATPase subunits as well as many additional genes involved in the lysosomal-autophagy pathway. Reduction of Mitf function leads to abnormal lysosomes and impairs autophagosome fusion and lipid breakdown during the response to starvation. In contrast, elevated Mitf levels increase the number of lysosomes, autophagosomes and autolysosomes, and decrease the size of lipid droplets. Inhibition of Drosophila MTORC1 induces Mitf translocation to the nucleus, underscoring conserved regulatory mechanisms between Drosophila and mammalian systems. Furthermore, we show Mitf-mediated clearance of cytosolic and nuclear expanded ATXN1 (ataxin 1) in a cellular model of spinocerebellar ataxia type 1 (SCA1). This remarkable observation illustrates the potential of the lysosomal-autophagy system to prevent toxic protein aggregation in both the cytoplasmic and nuclear compartments. We anticipate that the genetics of the Drosophila model and the absence of redundant MIT transcription factors will be exploited to investigate the regulation and function of the lysosomal-autophagy gene network.
Keywords: MTORC1; Mitf; TFEB; V-ATPase; autophagy; lipid metabolism; lysosome; proton pump.
Figures
References
-
- Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793: 664–73; PMID:18706940; http://dx.doi.org/10.1016/j.bbamcr.2008.07.014 - DOI - PubMed
-
- Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB 3rd. Autophagy: regulation and role in development. Autophagy 2013; 9:951–72; PMID:24121596; http://dx.doi.org/10.4161/auto.24273 - DOI - PMC - PubMed
-
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717–21; PMID:11099404; http://dx.doi.org/10.1126/science.290.5497.1717 - DOI - PMC - PubMed
-
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al.. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473–77; PMID:19556463 - PubMed
-
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429–33; PMID:21617040; http://dx.doi.org/10.1126/science.1204592 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
