MEP50/PRMT5 Reduces Gene Expression by Histone Arginine Methylation and this Is Reversed by PKCδ/p38δ Signaling
- PMID: 26763441
- PMCID: PMC4899982
- DOI: 10.1038/JID.2015.400
MEP50/PRMT5 Reduces Gene Expression by Histone Arginine Methylation and this Is Reversed by PKCδ/p38δ Signaling
Abstract
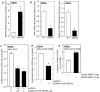
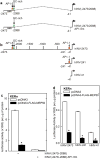
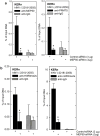
Protein kinase C δ (PKCδ) and p38δ are key proteins in a cascade that stimulates keratinocyte differentiation. This cascade activates transcription of involucrin (hINV) and other genes associated with differentiation. Protein arginine methyltransferase 5 (PRMT5) is an arginine methyltransferase that symmetrically dimethylates arginine residues. This protein interacts with a cofactor, methylosome protein 50 (MEP50), and symmetrically dimethylates arginine eight of histone 3 (H3R8me2s) and arginine three of histone 4 (H4R3me2s) to silence gene expression. We use the hINV gene as a tool to understand the relationship between PKCδ/p38δ and PRMT5/MEP50 signaling. MEP50 suppresses hINV mRNA level and promoter activity. This is associated with increased arginine dimethylation of hINV gene-associated H3/H4. We further show that the PKCδ/p38δ keratinocyte differentiation cascade reduces PRMT5 and MEP50 expression, association with the hINV gene promoter, and H3R8me2s and H4R2me2s formation. We propose that PRMT5/MEP50-dependent methylation is an epigenetic mechanism that assists in silencing of hINV expression, and that PKCδ signaling activates gene expression by directly activating transcription and by suppressing PRMT5/MEP50-dependent arginine dimethylation of promoter-associated histones. This is an example of crosstalk between PKCδ/p38δ signaling and PRMT5/MEP50 epigenetic silencing.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Methylosome Protein 50 and PKCδ/p38δ Protein Signaling Control Keratinocyte Proliferation via Opposing Effects on p21Cip1 Gene Expression.J Biol Chem. 2015 May 22;290(21):13521-30. doi: 10.1074/jbc.M115.642868. Epub 2015 Apr 7. J Biol Chem. 2015. PMID: 25851901 Free PMC article.
-
A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression.Oncogene. 2017 Jan 19;36(3):373-386. doi: 10.1038/onc.2016.205. Epub 2016 Jun 6. Oncogene. 2017. PMID: 27270440 Free PMC article.
-
Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase.J Biol Chem. 2015 Apr 10;290(15):9674-89. doi: 10.1074/jbc.M115.636894. Epub 2015 Feb 24. J Biol Chem. 2015. PMID: 25713080 Free PMC article.
-
The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond.Cell Mol Life Sci. 2015 Jun;72(11):2041-59. doi: 10.1007/s00018-015-1847-9. Epub 2015 Feb 7. Cell Mol Life Sci. 2015. PMID: 25662273 Free PMC article. Review.
-
Dismantling the epigenetic alliance: Emerging strategies to disrupt the PRMT5:MEP50 complex for cancer therapy.Eur J Med Chem. 2025 Oct 15;296:117870. doi: 10.1016/j.ejmech.2025.117870. Epub 2025 Jun 14. Eur J Med Chem. 2025. PMID: 40532498 Review.
Cited by
-
Protein Arginine Methyltransferase 5 Promotes pICln-Dependent Androgen Receptor Transcription in Castration-Resistant Prostate Cancer.Cancer Res. 2020 Nov 15;80(22):4904-4917. doi: 10.1158/0008-5472.CAN-20-1228. Epub 2020 Sep 30. Cancer Res. 2020. PMID: 32999000 Free PMC article.
-
Sulforaphane inhibits PRMT5 and MEP50 function to suppress the mesothelioma cancer cell phenotype.Mol Carcinog. 2021 Jul;60(7):429-439. doi: 10.1002/mc.23301. Epub 2021 Apr 19. Mol Carcinog. 2021. PMID: 33872411 Free PMC article.
-
Cellular consequences of arginine methylation.Cell Mol Life Sci. 2019 Aug;76(15):2933-2956. doi: 10.1007/s00018-019-03140-2. Epub 2019 May 17. Cell Mol Life Sci. 2019. PMID: 31101937 Free PMC article. Review.
-
The methylation induced by protein arginine methyltransferase 5 promotes tumorigenesis and progression of lung cancer.J Thorac Dis. 2018 Dec;10(12):7014-7019. doi: 10.21037/jtd.2018.10.100. J Thorac Dis. 2018. PMID: 30746248 Free PMC article. Review.
-
Arginine Methylation in Brain Tumors: Tumor Biology and Therapeutic Strategies.Cells. 2021 Jan 11;10(1):124. doi: 10.3390/cells10010124. Cells. 2021. PMID: 33440687 Free PMC article. Review.
References
-
- Adhikary G, Crish JF, Bone F, et al. An involucrin promoter AP1 transcription factor binding site is required for expression of involucrin in the corneal epithelium in vivo. Invest Ophthalmol Vis Sci. 2005;46:1219–1227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

