Inhibition of the FKBP family of peptidyl prolyl isomerases induces abortive translocation and degradation of the cellular prion protein
- PMID: 26764098
- PMCID: PMC4803302
- DOI: 10.1091/mbc.E15-10-0729
Inhibition of the FKBP family of peptidyl prolyl isomerases induces abortive translocation and degradation of the cellular prion protein
Abstract
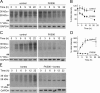
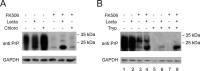
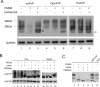
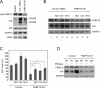
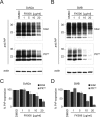
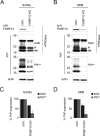
Prion diseases are fatal neurodegenerative disorders for which there is no effective treatment. Because the cellular prion protein (PrP(C)) is required for propagation of the infectious scrapie form of the protein, one therapeutic strategy is to reduce PrP(C) expression. Recently FK506, an inhibitor of the FKBP family of peptidyl prolyl isomerases, was shown to increase survival in animal models of prion disease, with proposed mechanisms including calcineurin inhibition, induction of autophagy, and reduced PrP(C) expression. We show that FK506 treatment results in a profound reduction in PrP(C) expression due to a defect in the translocation of PrP(C) into the endoplasmic reticulum with subsequent degradation by the proteasome. These phenotypes could be bypassed by replacing the PrP(C) signal sequence with that of prolactin or osteopontin. In mouse cells, depletion of ER luminal FKBP10 was almost as potent as FK506 in attenuating expression of PrP(C). However, this occurred at a later stage, after translocation of PrP(C) into the ER. Both FK506 treatment and FKBP10 depletion were effective in reducing PrP(Sc) propagation in cell models. These findings show the involvement of FKBP proteins at different stages of PrP(C) biogenesis and identify FKBP10 as a potential therapeutic target for the treatment of prion diseases.
© 2016 Stocki et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

References
-
- Barnes AM, Cabral WA, Weis M, Makareeva E, Mertz EL, Leikin S, Eyre D, Trujillo C, Marini JC. Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum Mutat. 2012;33:1589–1598. - PMC - PubMed
-
- Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. - PubMed
-
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

