FGFR and PTEN signaling interact during lens development to regulate cell survival
- PMID: 26764128
- PMCID: PMC4767529
- DOI: 10.1016/j.ydbio.2015.12.027
FGFR and PTEN signaling interact during lens development to regulate cell survival
Abstract
Lens epithelial cells express many receptor tyrosine kinases (RTKs) that stimulate PI3K-AKT and RAS-RAF-MEK-ERK intracellular signaling pathways. These pathways ultimately activate the phosphorylation of key cellular transcription factors and other proteins that control proliferation, survival, metabolism, and differentiation in virtually all cells. Among RTKs in the lens, only stimulation of fibroblast growth factor receptors (FGFRs) elicits a lens epithelial cell to fiber cell differentiation response in mammals. Moreover, although the lens expresses three different Fgfr genes, the isolated removal of Fgfr2 at the lens placode stage inhibits both lens cell survival and fiber cell differentiation. Phosphatase and tensin homolog (PTEN), commonly known as a tumor suppressor, inhibits ERK and AKT activation and initiates both apoptotic pathways, and cell cycle arrest. Here, we show that the combined deletion of Fgfr2 and Pten rescues the cell death phenotype associated with Fgfr2 loss alone. Additionally, Pten removal increased AKT and ERK activation, above the levels of controls, in the presence or absence of Fgfr2. However, isolated deletion of Pten failed to stimulate ectopic fiber cell differentiation, and the combined deletion of Pten and Fgfr2 failed to restore differentiation-specific Aquaporin0 and DnaseIIβ expression in the lens fiber cells.
Keywords: Differentiation; FGF Receptor; Lens; PTEN; Survival.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

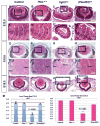


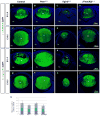


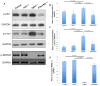
References
-
- Ahmad I, Iwata T, Leung HY. Mechanisms of FGFR-mediated carcinogenesis. Biochimica et biophysica acta. 2012;1823:850–860. - PubMed
-
- Bassnett S. Fiber cell denucleation in the primate lens. Investigative ophthalmology & visual science. 1997;38:1678–1687. - PubMed
-
- Bassnett S, Beebe DC. Lens Fiber Differentiation. In: Robinson ML, Lovicu FJ, editors. Development of the Ocular Lens. Cambridge; 2004. pp. 214–244.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

