A study of bias and increasing organismal complexity from their post-translational modifications and reaction site interplays
- PMID: 26764274
- PMCID: PMC5221421
- DOI: 10.1093/bib/bbv111
A study of bias and increasing organismal complexity from their post-translational modifications and reaction site interplays
Abstract
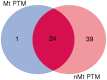
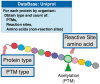
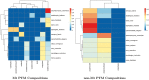
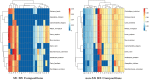
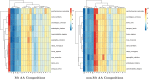
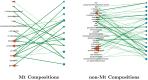
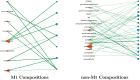
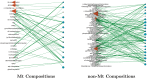
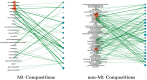
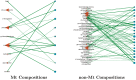
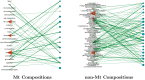
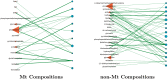
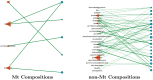
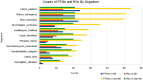
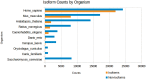
Post-translational modifications (PTMs) are important steps in the biosynthesis of proteins. Aside from their integral contributions to protein development, i.e. perform specialized proteolytic cleavage of regulatory subunits, the covalent addition of functional groups of proteins or the degradation of entire proteins, PTMs are also involved in enabling proteins to withstand and recover from temporary environmental stresses (heat shock, microgravity and many others). The literature supports evidence of thousands of recently discovered PTMs, many of which may likely contribute similarly (perhaps, even, interchangeably) to protein stress response. Although there are many PTM actors on the biological stage, our study determines that these PTMs are generally cast into organism-specific, preferential roles. In this work, we study the PTM compositions across the mitochondrial (Mt) and non-Mt proteomes of 11 diverse organisms to illustrate that each organism appears to have a unique list of PTMs, and an equally unique list of PTM-associated residue reaction sites (RSs), where PTMs interact with protein. Despite the present limitation of available PTM data across different species, we apply existing and current protein data to illustrate particular organismal biases. We explore the relative frequencies of observed PTMs, the RSs and general amino-acid compositions of Mt and non-Mt proteomes. We apply these data to create networks and heatmaps to illustrate the evidence of bias. We show that the number of PTMs and RSs appears to grow along with organismal complexity, which may imply that environmental stress could play a role in this bias.
Keywords: PTM bias; amino acid bias; organism complexity; reaction site bias.
© The Author 2016. Published by Oxford University Press. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Bonham-Carter O, Pedersen J, Najjar L, et al. Modeling the effects of microgravity on oxidation in mitochondria: a protein damage assessment across a diverse set of life forms. Comput Biol Med 2014;53:179–89. - PubMed
-
- Bonham-Carter O, Pedersen J, Najjar L, et al. Modeling the effects of microgravity on oxidation in mitochondria: a protein damage assessment across a diverse set of life forms. In: Data Mining Workshops (ICDMW), 2013 IEEE 13th International Conference on IEEE, 2013, pp. 250–7.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

