Promiscuous tumor targeting phage proteins
- PMID: 26764410
- PMCID: PMC4753993
- DOI: 10.1093/protein/gzv064
Promiscuous tumor targeting phage proteins
Abstract
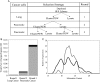
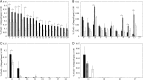
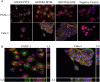
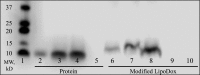
Cancer cell-specific targeting ligands against numerous cancer cell lines have been selected previously and used as ligands for cell-specific delivery of chemotherapies and various nanomedicines. However, tumor heterogeneity is one recognized problem hampering clinical translation of targeted anti-cancer medicines. Therefore, a novel class of targeting ligands is required that recognize receptors expressed between a variety of cancer phenotypes, identified here as 'promiscuous' ligands. In this work, promiscuous phage fusion proteins were first identified by a novel selection scheme to enrich for pan-cancer cell binding abilities, as indicated by conserved structural motifs identified previously in other cancer types. Additionally, peptide sequences containing a combination of motifs were identified to modulate binding. A panel of phage fusion proteins was studied for their specificity and selectivity for lung and pancreatic cancer cells. Phage displaying the fusion peptides GSLEEVSTL or GEFDELMTM, the two predominate clones with greatest binding ability, were used to modify preformed, doxorubicin-loaded, liposomes. These modified liposomes increased cytotoxicity up to 8.1-fold in several cancer cell lines when compared with unmodified liposomal doxorubicin. Taken together, these data indicate that promiscuous phage proteins, selected against different cancer cell lines, can be used as targeting ligands for treatment of heterogeneous tumor populations.
Keywords: lung cancer; multi-target selection; pancreatic cancer; phage display; targeted nanomedicines.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Selection of pancreatic cancer cell-binding landscape phages and their use in development of anticancer nanomedicines.Protein Eng Des Sel. 2014 Jul;27(7):235-43. doi: 10.1093/protein/gzu020. Epub 2014 Jun 4. Protein Eng Des Sel. 2014. PMID: 24899628 Free PMC article.
-
Landscape phage fusion protein-mediated targeting of nanomedicines enhances their prostate tumor cell association and cytotoxic efficiency.Nanomedicine. 2010 Aug;6(4):538-46. doi: 10.1016/j.nano.2010.01.005. Epub 2010 Feb 4. Nanomedicine. 2010. PMID: 20138246 Free PMC article.
-
Landscape phages and their fusion proteins targeted to breast cancer cells.Protein Eng Des Sel. 2012 Jun;25(6):271-83. doi: 10.1093/protein/gzs013. Epub 2012 Apr 6. Protein Eng Des Sel. 2012. PMID: 22490956 Free PMC article.
-
Phage display biopanning and isolation of target-unrelated peptides: in search of nonspecific binders hidden in a combinatorial library.Amino Acids. 2016 Dec;48(12):2699-2716. doi: 10.1007/s00726-016-2329-6. Epub 2016 Sep 20. Amino Acids. 2016. PMID: 27650972 Review.
-
Peptide functionalized liposomes for receptor targeted cancer therapy.APL Bioeng. 2021 Jan 25;5(1):011501. doi: 10.1063/5.0029860. eCollection 2021 Mar. APL Bioeng. 2021. PMID: 33532673 Free PMC article. Review.
Cited by
-
Combinatorial Avidity Selection of Mosaic Landscape Phages Targeted at Breast Cancer Cells-An Alternative Mechanism of Directed Molecular Evolution.Viruses. 2019 Aug 26;11(9):785. doi: 10.3390/v11090785. Viruses. 2019. PMID: 31454976 Free PMC article.
-
Virus-Derived Peptides for Clinical Applications.Chem Rev. 2017 Aug 9;117(15):10377-10402. doi: 10.1021/acs.chemrev.7b00100. Epub 2017 Jul 19. Chem Rev. 2017. PMID: 28723101 Free PMC article. Review.
-
Paradigm shift in bacteriophage-mediated delivery of anticancer drugs: from targeted 'magic bullets' to self-navigated 'magic missiles'.Expert Opin Drug Deliv. 2017 Mar;14(3):373-384. doi: 10.1080/17425247.2016.1218463. Epub 2016 Aug 5. Expert Opin Drug Deliv. 2017. PMID: 27466706 Free PMC article. Review.
-
Landscape Phage: Evolution from Phage Display to Nanobiotechnology.Viruses. 2018 Jun 7;10(6):311. doi: 10.3390/v10060311. Viruses. 2018. PMID: 29880747 Free PMC article. Review.
-
Phage-Displayed Mimotopes of SARS-CoV-2 Spike Protein Targeted to Authentic and Alternative Cellular Receptors.Viruses. 2022 Feb 14;14(2):384. doi: 10.3390/v14020384. Viruses. 2022. PMID: 35215976 Free PMC article.
References
-
- Arap W., Pasqualini R., Ruoslahti E. (1998) Science, 279, 377–380. - PubMed
-
- Brigati J.R., Petrenko V.A. (2005) Anal. Bioanal. Chem., 382, 1346–1350. - PubMed
-
- Brigati J.R., Samoylova T.I., Jayanna P.K., Petrenko V.A. (2008) Phage display for generating peptide reagents. Current protocols in protein science/editorial board, John E Coligan [et al],Chapter 18, Unit 18.19. First published on 2008/04/23, doi: 10.1002/0471140864.ps1809s51.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

