The Host Shapes the Gut Microbiota via Fecal MicroRNA
- PMID: 26764595
- PMCID: PMC4847146
- DOI: 10.1016/j.chom.2015.12.005
The Host Shapes the Gut Microbiota via Fecal MicroRNA
Abstract
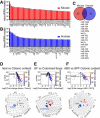
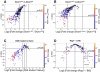
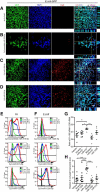
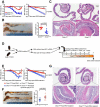
The host gut microbiota varies across species and individuals but is relatively stable over time within an individual. How the host selectively shapes the microbiota is largely unclear. Here, we show that fecal microRNA (miRNA)-mediated inter-species gene regulation facilitates host control of the gut microbiota. miRNAs are abundant in mouse and human fecal samples and present within extracellular vesicles. Cell-specific loss of the miRNA-processing enzyme, Dicer, identified intestinal epithelial cells (IEC) and Hopx-positive cells as predominant fecal miRNA sources. These miRNAs can enter bacteria, such as F. nucleatum and E. coli, specifically regulate bacterial gene transcripts, and affect bacterial growth. IEC-miRNA-deficient (Dicer1(ΔIEC)) mice exhibit uncontrolled gut microbiota and exacerbated colitis, and WT fecal miRNA transplantation restores fecal microbes and ameliorates colitis. These findings identify both a physiologic role by which fecal miRNA shapes the gut microbiota and a potential strategy for manipulating the microbiome.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Gut microbiota: Host faecal miRNA regulates gut microbiota.Nat Rev Gastroenterol Hepatol. 2016 Mar;13(3):122-3. doi: 10.1038/nrgastro.2016.19. Epub 2016 Feb 3. Nat Rev Gastroenterol Hepatol. 2016. PMID: 26837709 No abstract available.
-
Control of the gut microbiome by fecal microRNA.Microb Cell. 2016 Mar 9;3(4):176-177. doi: 10.15698/mic2016.04.492. Microb Cell. 2016. PMID: 28357349 Free PMC article.
Similar articles
-
Control of the gut microbiome by fecal microRNA.Microb Cell. 2016 Mar 9;3(4):176-177. doi: 10.15698/mic2016.04.492. Microb Cell. 2016. PMID: 28357349 Free PMC article.
-
Spatial heterogeneity of bacterial colonization across different gut segments following inter-species microbiota transplantation.Microbiome. 2020 Nov 18;8(1):161. doi: 10.1186/s40168-020-00917-7. Microbiome. 2020. PMID: 33208178 Free PMC article.
-
Human-derived microRNA 21 regulates indole and L-tryptophan biosynthesis transcripts in the gut commensal Bacteroides thetaiotaomicron.mBio. 2025 Mar 12;16(3):e0392824. doi: 10.1128/mbio.03928-24. Epub 2025 Jan 29. mBio. 2025. PMID: 39878512 Free PMC article.
-
[MicroRNA regulation on host-microbiota interaction--a review].Wei Sheng Wu Xue Bao. 2013 Oct 4;53(10):1018-24. Wei Sheng Wu Xue Bao. 2013. PMID: 24409756 Review. Chinese.
-
[Advances in host-microbe metabolic axis].Wei Sheng Wu Xue Bao. 2017 Feb 4;57(2):161-9. Wei Sheng Wu Xue Bao. 2017. PMID: 29750478 Review. Chinese.
Cited by
-
An In Vitro System Mimics the Intestinal Microbiota of Striped Beakfish (Oplegnathus fasciatus) and Inhibits Vibrio alginolyticus by Limosilactobacillus reuteri-Derived Extracellular Vesicles.Animals (Basel). 2024 Jun 14;14(12):1792. doi: 10.3390/ani14121792. Animals (Basel). 2024. PMID: 38929411 Free PMC article.
-
miRNA Profiling in the Chicken Liver under the Influence of Early Microbiota Stimulation with Probiotic, Prebiotic, and Synbiotic.Genes (Basel). 2021 May 1;12(5):685. doi: 10.3390/genes12050685. Genes (Basel). 2021. PMID: 34062867 Free PMC article.
-
Colon Cancer and Obesity: A Narrative Review.Cureus. 2022 Aug 1;14(8):e27589. doi: 10.7759/cureus.27589. eCollection 2022 Aug. Cureus. 2022. PMID: 36059323 Free PMC article. Review.
-
Evolution, the Immune System, and the Health Consequences of Socioeconomic Inequality.mSystems. 2022 Apr 26;7(2):e0143821. doi: 10.1128/msystems.01438-21. Epub 2022 Mar 14. mSystems. 2022. PMID: 35285679 Free PMC article.
-
Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota.Cell Host Microbe. 2018 Nov 14;24(5):637-652.e8. doi: 10.1016/j.chom.2018.10.001. Epub 2018 Oct 25. Cell Host Microbe. 2018. PMID: 30449315 Free PMC article.
References
-
- Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–295. - PubMed
-
- Banerjee RR, Lazar MA. Dimerization of resistin and resistin-like molecules is determined by a single cysteine. J. Biol. Chem. 2001;276:25970–25973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

