Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori
- PMID: 26764597
- PMCID: PMC4793151
- DOI: 10.1016/j.chom.2015.12.004
Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori
Abstract
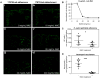
The Helicobacter pylori adhesin BabA binds mucosal ABO/Le(b) blood group (bg) carbohydrates. BabA facilitates bacterial attachment to gastric surfaces, increasing strain virulence and forming a recognized risk factor for peptic ulcers and gastric cancer. High sequence variation causes BabA functional diversity, but the underlying structural-molecular determinants are unknown. We generated X-ray structures of representative BabA isoforms that reveal a polymorphic, three-pronged Le(b) binding site. Two diversity loops, DL1 and DL2, provide adaptive control to binding affinity, notably ABO versus O bg preference. H. pylori strains can switch bg preference with single DL1 amino acid substitutions, and can coexpress functionally divergent BabA isoforms. The anchor point for receptor binding is the embrace of an ABO fucose residue by a disulfide-clasped loop, which is inactivated by reduction. Treatment with the redox-active pharmaceutic N-acetylcysteine lowers gastric mucosal neutrophil infiltration in H. pylori-infected Le(b)-expressing mice, providing perspectives on possible H. pylori eradication therapies.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
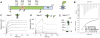





Comment in
-
H. pylori's BabA Embraces Change.Cell Host Microbe. 2016 Jan 13;19(1):5-7. doi: 10.1016/j.chom.2015.12.013. Cell Host Microbe. 2016. PMID: 26764590
Similar articles
-
Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans.mBio. 2014 Dec 16;5(6):e02281-14. doi: 10.1128/mBio.02281-14. mBio. 2014. PMID: 25516619 Free PMC article.
-
Helicobacter pylori Strains from Duodenal Ulcer Patients Exhibit Mixed babA/B Genotypes with Low Levels of BabA Adhesin and Lewis b Binding.Dig Dis Sci. 2016 Oct;61(10):2868-2877. doi: 10.1007/s10620-016-4217-z. Epub 2016 Jun 18. Dig Dis Sci. 2016. PMID: 27318698
-
Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin.Science. 2004 Jul 23;305(5683):519-22. doi: 10.1126/science.1098801. Science. 2004. PMID: 15273394
-
Histo-blood group carbohydrates as facilitators for infection by Helicobacter pylori.Infect Genet Evol. 2017 Sep;53:167-174. doi: 10.1016/j.meegid.2017.05.025. Epub 2017 May 31. Infect Genet Evol. 2017. PMID: 28577915 Review.
-
Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis.World J Gastroenterol. 2008 Jul 21;14(27):4265-72. doi: 10.3748/wjg.14.4265. World J Gastroenterol. 2008. PMID: 18666312 Free PMC article. Review.
Cited by
-
Helicobacter pylori Outer Membrane Protein-Related Pathogenesis.Toxins (Basel). 2017 Mar 11;9(3):101. doi: 10.3390/toxins9030101. Toxins (Basel). 2017. PMID: 28287480 Free PMC article. Review.
-
Multiple surface interaction mechanisms direct the anchoring, co-aggregation and formation of dual-species biofilm between Candida albicans and Helicobacter pylori.J Adv Res. 2021 Mar 31;35:169-185. doi: 10.1016/j.jare.2021.03.013. eCollection 2022 Jan. J Adv Res. 2021. PMID: 35024198 Free PMC article.
-
Helicobacter pylori adhesin HopQ disrupts trans dimerization in human CEACAMs.EMBO J. 2018 Jul 2;37(13):e98665. doi: 10.15252/embj.201798665. Epub 2018 Jun 1. EMBO J. 2018. PMID: 29858229 Free PMC article.
-
Helicobacter pylori SabA binding gangliosides of human stomach.Virulence. 2018 Dec 31;9(1):738-751. doi: 10.1080/21505594.2018.1440171. Virulence. 2018. PMID: 29473478 Free PMC article.
-
Helicobacter pylori in Human Stomach: The Inconsistencies in Clinical Outcomes and the Probable Causes.Front Microbiol. 2021 Aug 17;12:713955. doi: 10.3389/fmicb.2021.713955. eCollection 2021. Front Microbiol. 2021. PMID: 34484153 Free PMC article. Review.
References
-
- Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A, et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. - PubMed
-
- Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–35271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

