Replication of an Autonomous Human Parvovirus in Non-dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways
- PMID: 26765330
- PMCID: PMC4713420
- DOI: 10.1371/journal.ppat.1005399
Replication of an Autonomous Human Parvovirus in Non-dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways
Abstract
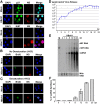
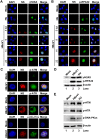
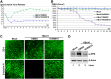
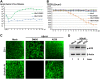
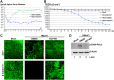
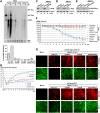
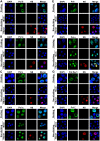
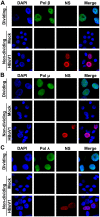
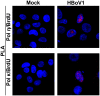
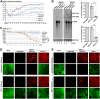
Human bocavirus 1 (HBoV1) belongs to the genus Bocaparvovirus of the Parvoviridae family, and is an emerging human pathogenic respiratory virus. In vitro, HBoV1 infects well-differentiated/polarized primary human airway epithelium (HAE) cultured at an air-liquid interface (HAE-ALI). Although it is well known that autonomous parvovirus replication depends on the S phase of the host cells, we demonstrate here that the HBoV1 genome amplifies efficiently in mitotically quiescent airway epithelial cells of HAE-ALI cultures. Analysis of HBoV1 DNA in infected HAE-ALI revealed that HBoV1 amplifies its ssDNA genome following a typical parvovirus rolling-hairpin DNA replication mechanism. Notably, HBoV1 infection of HAE-ALI initiates a DNA damage response (DDR) with activation of all three phosphatidylinositol 3-kinase-related kinases (PI3KKs). We found that the activation of the three PI3KKs is required for HBoV1 genome amplification; and, more importantly, we identified that two Y-family DNA polymerases, Pol η and Pol κ, are involved in HBoV1 genome amplification. Overall, we have provided an example of de novo DNA synthesis (genome amplification) of an autonomous parvovirus in non-dividing cells, which is dependent on the cellular DNA damage and repair pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
DNA Damage Signaling Is Required for Replication of Human Bocavirus 1 DNA in Dividing HEK293 Cells.J Virol. 2016 Dec 16;91(1):e01831-16. doi: 10.1128/JVI.01831-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27733644 Free PMC article.
-
The Large Nonstructural Protein (NS1) of Human Bocavirus 1 Directly Interacts with Ku70, Which Plays an Important Role in Virus Replication in Human Airway Epithelia.J Virol. 2022 Feb 23;96(4):e0184021. doi: 10.1128/JVI.01840-21. Epub 2021 Dec 8. J Virol. 2022. PMID: 34878919 Free PMC article.
-
Identification and Functional Analysis of Novel Nonstructural Proteins of Human Bocavirus 1.J Virol. 2015 Oct;89(19):10097-109. doi: 10.1128/JVI.01374-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223640 Free PMC article.
-
Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications.Front Microbiol. 2021 Jun 16;12:696604. doi: 10.3389/fmicb.2021.696604. eCollection 2021. Front Microbiol. 2021. PMID: 34220786 Free PMC article. Review.
-
[Progress on development and research of human bocavirus 1].Bing Du Xue Bao. 2014 Jan;30(1):103-8. Bing Du Xue Bao. 2014. PMID: 24772907 Review. Chinese.
Cited by
-
Mutations in the C-terminus of HBoV NS1 affect the function of NP1.Sci Rep. 2017 Aug 7;7(1):7407. doi: 10.1038/s41598-017-06513-4. Sci Rep. 2017. PMID: 28785044 Free PMC article.
-
Endonuclease Activity Inhibition of the NS1 Protein of Parvovirus B19 as a Novel Target for Antiviral Drug Development.Antimicrob Agents Chemother. 2019 Feb 26;63(3):e01879-18. doi: 10.1128/AAC.01879-18. Print 2019 Mar. Antimicrob Agents Chemother. 2019. PMID: 30530599 Free PMC article.
-
Screening out microRNAs and Their Molecular Pathways with a Potential Role in the Regulation of Parvovirus B19 Infection Through In Silico Analysis.Int J Mol Sci. 2025 May 23;26(11):5038. doi: 10.3390/ijms26115038. Int J Mol Sci. 2025. PMID: 40507847 Free PMC article.
-
Human Bocavirus Infection of Permanent Cells Differentiated to Air-Liquid Interface Cultures Activates Transcription of Pathways Involved in Tumorigenesis.Cancers (Basel). 2018 Oct 30;10(11):410. doi: 10.3390/cancers10110410. Cancers (Basel). 2018. PMID: 30380741 Free PMC article.
-
RNA Binding Motif Protein RBM45 Regulates Expression of the 11-Kilodalton Protein of Parvovirus B19 through Binding to Novel Intron Splicing Enhancers.mBio. 2020 Mar 10;11(2):e00192-20. doi: 10.1128/mBio.00192-20. mBio. 2020. PMID: 32156816 Free PMC article.
References
-
- Meriluoto M, Hedman L, Tanner L, Simell V, Makinen M, Simell S, Mykkanen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Soderlund-Venermo M (2012) Association of Human Bocavirus 1 Infection with Respiratory Disease in Childhood Follow-up Study, Finland. Emerg Infect Dis 18: 264–271. 10.3201/eid1802.111293 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

