The Interaction of Myc with Miz1 Defines Medulloblastoma Subgroup Identity
- PMID: 26766587
- PMCID: PMC4714043
- DOI: 10.1016/j.ccell.2015.12.003
The Interaction of Myc with Miz1 Defines Medulloblastoma Subgroup Identity
Abstract
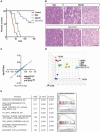
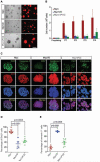
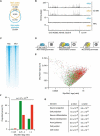
Four distinct subgroups of cerebellar medulloblastomas (MBs) differ in their histopathology, molecular profiles, and prognosis. c-Myc (Myc) or MycN overexpression in granule neuron progenitors (GNPs) induces Group 3 (G3) or Sonic Hedgehog (SHH) MBs, respectively. Differences in Myc and MycN transcriptional profiles depend, in part, on their interaction with Miz1, which binds strongly to Myc but not MycN, to target sites on chromatin. Myc suppresses ciliogenesis and reprograms the transcriptome of SHH-dependent GNPs through Miz1-dependent gene repression to maintain stemness. Genetic disruption of the Myc/Miz1 interaction inhibited G3 MB development. Target genes of Myc/Miz1 are repressed in human G3 MBs but not in other subgroups. Therefore, the Myc/Miz1 interaction is a defining hallmark of G3 MB development.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Parsing Myc Paralogs in Oncogenesis.Cancer Cell. 2016 Jan 11;29(1):1-2. doi: 10.1016/j.ccell.2015.12.009. Cancer Cell. 2016. PMID: 26766585
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
