An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma
- PMID: 26766592
- PMCID: PMC4809063
- DOI: 10.1016/j.ccell.2015.12.004
An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma
Abstract
Dysregulated metabolism is a hallmark of cancer, manifested through alterations in metabolites. We performed metabolomic profiling on 138 matched clear cell renal cell carcinoma (ccRCC)/normal tissue pairs and found that ccRCC is characterized by broad shifts in central carbon metabolism, one-carbon metabolism, and antioxidant response. Tumor progression and metastasis were associated with metabolite increases in glutathione and cysteine/methionine metabolism pathways. We develop an analytic pipeline and visualization tool (metabolograms) to bridge the gap between TCGA transcriptomic profiling and our metabolomic data, which enables us to assemble an integrated pathway-level metabolic atlas and to demonstrate discordance between transcriptome and metabolome. Lastly, expression profiling was performed on a high-glutathione cluster, which corresponds to a poor-survival subgroup in the ccRCC TCGA cohort.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
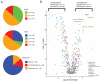


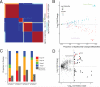

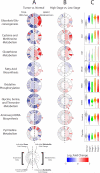
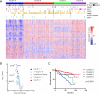
Comment in
-
Kidney cancer: When worlds collide--integrating metabolic and transcriptomic data in ccRCC.Nat Rev Urol. 2016 Mar;13(3):126. doi: 10.1038/nrurol.2016.16. Epub 2016 Jan 27. Nat Rev Urol. 2016. PMID: 26813954 No abstract available.
-
Multi-omics approach reveals the secrets of metabolism of clear cell-renal cell carcinoma.Transl Androl Urol. 2016 Oct;5(5):801-803. doi: 10.21037/tau.2016.06.12. Transl Androl Urol. 2016. PMID: 27785441 Free PMC article. No abstract available.
-
Commentary on: "An integrated metabolic atlas of clear cell renal cell carcinoma." Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, Chen Y, Stirdivant SM, Russo P, Chen YB, Tickoo SK, Reuter VE, Cheng EH, Sander C, Hsieh JJ.: Cancer Cell. 2016 Jan 11;29(1):104-16.Urol Oncol. 2017 Sep;35(9):579-580. doi: 10.1016/j.urolonc.2017.07.023. Epub 2017 Aug 5. Urol Oncol. 2017. PMID: 28789930
References
-
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature protocols. 2013;8:1765–1786. - PubMed
-
- Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, Karoly E, Qin LX, Ye QH, Jia HL, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075. e1061. - PMC - PubMed
-
- Casero RA, Jr., Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature reviews Drug discovery. 2007;6:373–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
