Complex regulation of ADAR-mediated RNA-editing across tissues
- PMID: 26768488
- PMCID: PMC4714477
- DOI: 10.1186/s12864-015-2291-9
Complex regulation of ADAR-mediated RNA-editing across tissues
Abstract
Background: RNA-editing is a tightly regulated, and essential cellular process for a properly functioning brain. Dysfunction of A-to-I RNA editing can have catastrophic effects, particularly in the central nervous system. Thus, understanding how the process of RNA-editing is regulated has important implications for human health. However, at present, very little is known about the regulation of editing across tissues, and individuals.
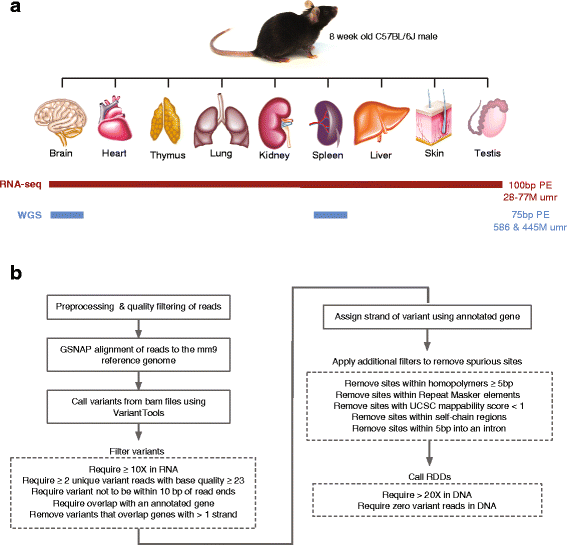
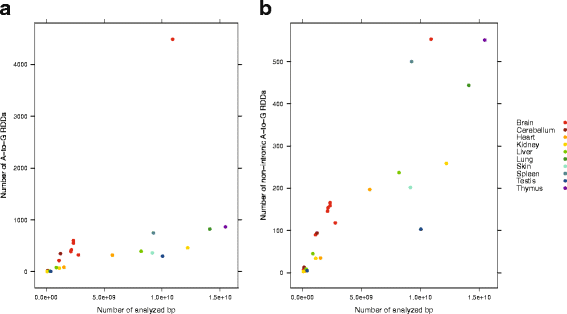
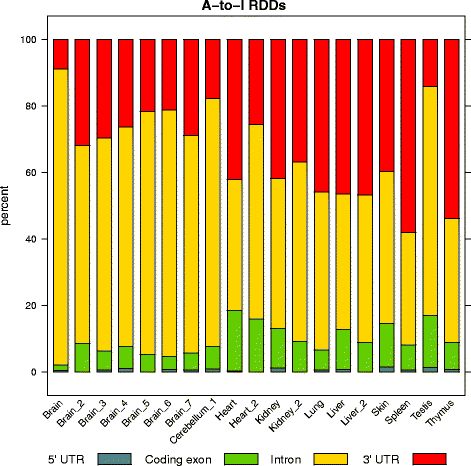
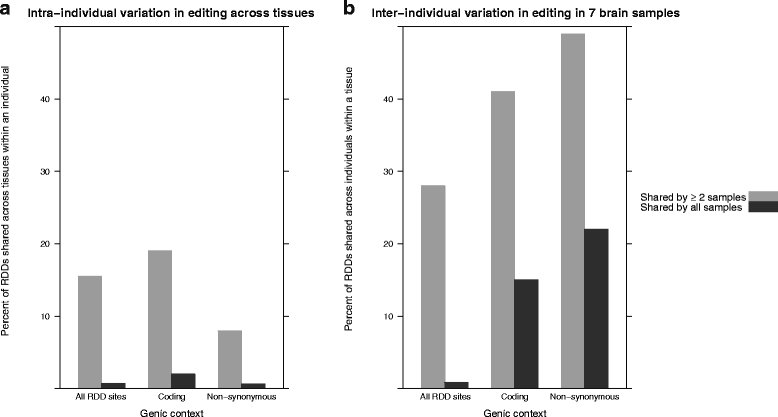
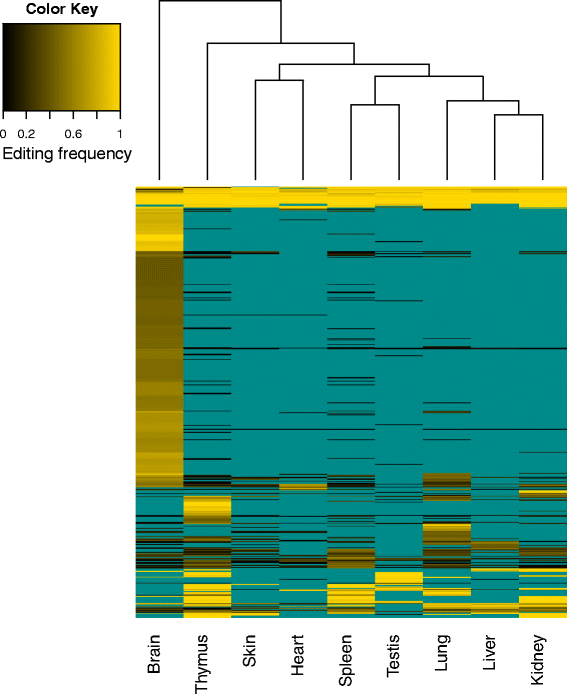
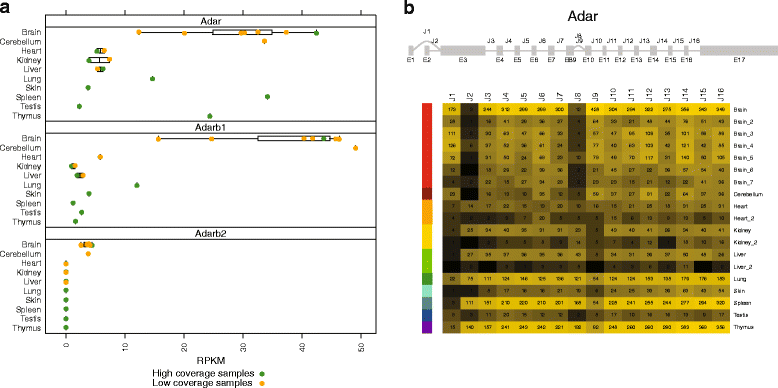
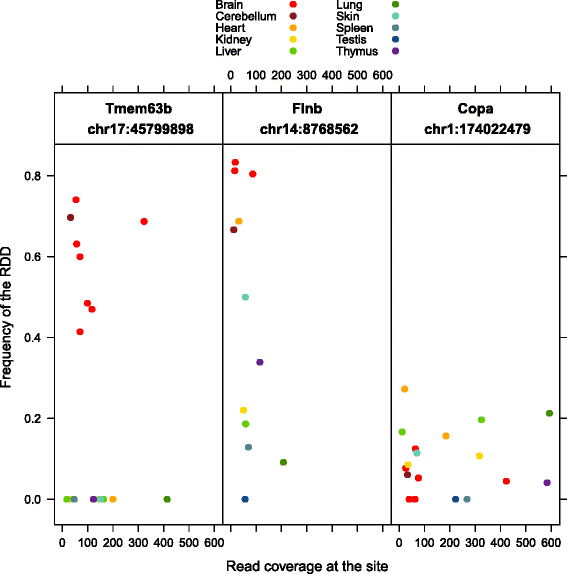
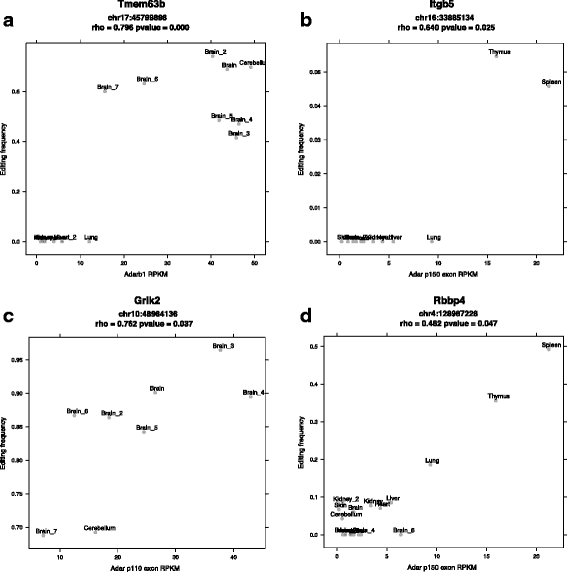
Results: Here we present an analysis of RNA-editing patterns from 9 different tissues harvested from a single mouse. For comparison, we also analyzed data for 5 of these tissues harvested from 15 additional animals. We find that tissue specificity of editing largely reflects differential expression of substrate transcripts across tissues. We identified a surprising enrichment of editing in intronic regions of brain transcripts, that could account for previously reported higher levels of editing in brain. There exists a small but remarkable amount of editing which is tissue-specific, despite comparable expression levels of the edit site across multiple tissues. Expression levels of editing enzymes and their isoforms can explain some, but not all of this variation.
Conclusions: Together, these data suggest a complex regulation of the RNA-editing process beyond transcript expression levels.
Figures
References
-
- Polson AG, Crain PF, Pomerantz SC, McCloskey JA, Bass BL. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991;30:11507–14. doi: 10.1021/bi00113a004. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

