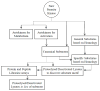
Enzyme Activity Assays for Protein Kinases: Strategies to Identify Active Substrates
- PMID: 26768716
- PMCID: PMC5599267
- DOI: 10.2174/1570163813666160115125930
Enzyme Activity Assays for Protein Kinases: Strategies to Identify Active Substrates
Abstract
Protein kinases are an important class of enzymes and drug targets. New opportunities to discover medicines for neglected diseases can be leveraged by the extensive kinase tools and knowledge created in targeting human kinases. A valuable tool for kinase drug discovery is an enzyme assay that measures catalytic function. The functional assay can be used to identify inhibitors, estimate affinity, characterize molecular mechanisms of action (MMOAs) and evaluate selectivity. However, establishing an enzyme assay for a new kinases requires identification of a suitable substrate. Identification of a new kinase's endogenous physiologic substrate and function can be extremely costly and time consuming. Fortunately, most kinases are promiscuous and will catalyze the phosphotransfer from ATP to alternative substrates with differing degrees of catalytic efficiency. In this manuscript we review strategies and successes in the identification of alternative substrates for kinases from organisms responsible for many of the neglected tropical diseases (NTDs) towards the goal of informing strategies to identify substrates for new kinases. Approaches for establishing a functional kinase assay include measuring auto-activation and use of generic substrates and peptides. The most commonly used generic substrates are casein, myelin basic protein, and histone. Sequence homology modeling can provide insights into the potential substrates and the requirement for activation. Empirical approaches that can identify substrates include screening of lysates (which may also help identify native substrates) and use of peptide arrays. All of these approaches have been used with a varying degree of success to identify alternative substrates.
Conflict of interest statement
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
Similar articles
-
Protein kinase substrate identification on functional protein arrays.BMC Biotechnol. 2008 Feb 28;8:22. doi: 10.1186/1472-6750-8-22. BMC Biotechnol. 2008. PMID: 18307815 Free PMC article.
-
Protein kinase structure and function analysis with chemical tools.Biochim Biophys Acta. 2005 Dec 30;1754(1-2):65-78. doi: 10.1016/j.bbapap.2005.08.020. Epub 2005 Sep 13. Biochim Biophys Acta. 2005. PMID: 16213197 Review.
-
Simultaneously synthesized peptides on continuous cellulose membranes as substrates for protein kinases.Pept Res. 1996 Jan-Feb;9(1):6-11. Pept Res. 1996. PMID: 8727478
-
High-throughput kinase assays with protein substrates using fluorescent polymer superquenching.BMC Biotechnol. 2005 May 31;5:16. doi: 10.1186/1472-6750-5-16. BMC Biotechnol. 2005. PMID: 15927069 Free PMC article.
-
Identification of protein kinase substrates by proteomic approaches.Expert Rev Proteomics. 2008 Jun;5(3):497-505. doi: 10.1586/14789450.5.3.497. Expert Rev Proteomics. 2008. PMID: 18532915 Review.
Cited by
-
The Legionella Effector Kinase LegK7 Hijacks the Host Hippo Pathway to Promote Infection.Cell Host Microbe. 2018 Sep 12;24(3):429-438.e6. doi: 10.1016/j.chom.2018.08.004. Cell Host Microbe. 2018. PMID: 30212651 Free PMC article.
-
System-Wide Profiling of Ubiquitin E3 Ligase Activities with Quantitative Proteomics and Bioinformatics Analysis.Methods Mol Biol. 2025;2921:345-359. doi: 10.1007/978-1-0716-4502-4_19. Methods Mol Biol. 2025. PMID: 40516000
-
From the research laboratory to the database: the Caenorhabditis elegans kinome in UniProtKB.Biochem J. 2017 Feb 15;474(4):493-515. doi: 10.1042/BCJ20160991. Biochem J. 2017. PMID: 28159896 Free PMC article. Review.
-
A Radioactive in vitro ERK3 Kinase Assay.Bio Protoc. 2019 Aug 20;9(16):e3332. doi: 10.21769/BioProtoc.3332. Bio Protoc. 2019. PMID: 31930160 Free PMC article.
-
Proteomic approaches for protein kinase substrate identification in Apicomplexa.Mol Biochem Parasitol. 2024 Sep;259:111633. doi: 10.1016/j.molbiopara.2024.111633. Epub 2024 May 29. Mol Biochem Parasitol. 2024. PMID: 38821187 Review.
References
-
- Sawyer TK, Wu JC, Sawyer JR, English JM. Protein kinase inhibitors: breakthrough medicines and the next generation. Expert Opin Investig Drugs. 2013;22(6):675–8. - PubMed
-
- Simmons DL. Targeting kinases: a new approach to treating inflammatory rheumatic diseases. Curr Opin Pharmacol. 2013;13(3):426–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources