Activating the branch-forming splicing pathway by reengineering the ribozyme component of a natural group II intron
- PMID: 26769855
- PMCID: PMC4748821
- DOI: 10.1261/rna.054643.115
Activating the branch-forming splicing pathway by reengineering the ribozyme component of a natural group II intron
Abstract
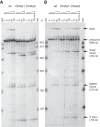
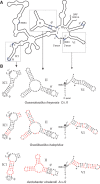
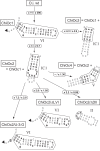
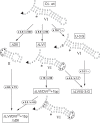
When assayed in vitro, group IIC self-splicing introns, which target bacterial Rho-independent transcription terminators, generally fail to yield branched products during splicing despite their possessing a seemingly normal branchpoint. Starting with intron O.i.I1 from Oceanobacillus iheyensis, whose crystallographically determined structure lacks branchpoint-containing domain VI, we attempted to determine what makes this intron unfit for in vitro branch formation. A major factor was found to be the length of the helix at the base of domain VI: 4 base pairs (bp) are required for efficient branching, even though a majority of group IIC introns have a 3-bp helix. Equally important for lariat formation is the removal of interactions between ribozyme domains II and VI, which are specific to the second step of splicing. Conversely, mismatching of domain VI and its proposed first-step receptor in subdomain IC1 was found to be detrimental; these data suggest that the intron-encoded protein may promote branch formation partly by modulating the equilibrium between conformations specific to the first and second steps of splicing. As a practical application, we show that by making just two changes to the O.i.I1 ribozyme, it is possible to generate sufficient amounts of lariat intron for the latter to be purified and used in kinetic assays in which folding and reaction are uncoupled.
Keywords: group II intron; lariat intron; linear intron; self-splicing.
© 2016 Monachello et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Adamidi C, Fedorova O, Pyle AM. 2003. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42: 3409–3418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
