16S rRNA gene sequencing is a non-culture method of defining the specific bacterial etiology of ventilator-associated pneumonia
- PMID: 26770469
- PMCID: PMC4694369
16S rRNA gene sequencing is a non-culture method of defining the specific bacterial etiology of ventilator-associated pneumonia
Abstract
Ventilator-associated pneumonia (VAP) is an acquired respiratory tract infection following tracheal intubation. The most common hospital-acquired infection among patients with acute respiratory failure, VAP is associated with a mortality rate of 20-30%. The standard bacterial culture method for identifying the etiology of VAP is not specific, timely, or accurate in identifying the bacterial pathogens. This study used 16S rRNA gene metagenomic sequencing to identify and quantify the pathogenic bacteria in lower respiratory tract and oropharyngeal samples of 55 VAP patients. Sequencing of the 16S rRNA gene has served as a valuable tool in bacterial identification, particularly when other biochemical, molecular, or phenotypic identification techniques fail. In this study, 16S rRNA gene sequencing was performed in parallel with the standard bacterial culture method to identify and quantify bacteria present in the collected patient samples. Sequence analysis showed the colonization of multidrug-resistant strains in VAP secretions. Further, this method identified Prevotella, Proteus, Aquabacter, and Sphingomonas bacterial genera that were not detected by the standard bacterial culture method. Seven categories of bacteria, Streptococcus, Neisseria, Corynebacterium, Acinetobacter, Staphylococcus, Pseudomonas and Klebsiella, were detectable by both 16S rRNA gene sequencing and standard bacterial culture methods. Further, 16S rRNA gene sequencing had a significantly higher sensitivity in detecting Streptococcus and Pseudomonas when compared to standard bacterial culture. Together, these data present 16S rRNA gene sequencing as a novel VAP diagnosis tool that will further enable pathogen-specific treatment of VAP.
Keywords: 16S rDNA; 16S rRNA gene sequencing; Ventilator-associated pneumonia (VAP); lower respiratory tract; oropharynx.
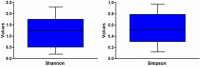
Figures
Similar articles
-
[The use of 16S rDNA sequencing in species diversity analysis for sputum of patients with ventilator-associated pneumonia].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2014 May;26(5):294-9. doi: 10.3760/cma.j.issn.2095-4352.2014.05.002. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2014. PMID: 24809255 Chinese.
-
Identification of respiratory microbiota markers in ventilator-associated pneumonia.Intensive Care Med. 2019 Aug;45(8):1082-1092. doi: 10.1007/s00134-019-05660-8. Epub 2019 Jun 17. Intensive Care Med. 2019. PMID: 31209523 Free PMC article.
-
High-throughput sequencing of 16S rDNA amplicons characterizes bacterial composition in bronchoalveolar lavage fluid in patients with ventilator-associated pneumonia.Drug Des Devel Ther. 2015 Aug 18;9:4883-96. doi: 10.2147/DDDT.S87634. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26345636 Free PMC article.
-
The microbiology of ventilator-associated pneumonia.Respir Care. 2005 Jun;50(6):742-63; discussion 763-5. Respir Care. 2005. PMID: 15913466 Review.
-
Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.Clin Microbiol Infect. 2008 Oct;14(10):908-34. doi: 10.1111/j.1469-0691.2008.02070.x. Clin Microbiol Infect. 2008. PMID: 18828852 Review.
Cited by
-
Respiratory microbiome profiles differ by recent hospitalization and nursing home residence in patients on mechanical ventilation.J Transl Med. 2020 Dec 7;18(1):464. doi: 10.1186/s12967-020-02642-z. J Transl Med. 2020. PMID: 33287847 Free PMC article.
-
Clinical characteristics and drug susceptibility patterns of Corynebacterium species in bacteremic patients with hematological disorders.Eur J Clin Microbiol Infect Dis. 2021 Oct;40(10):2095-2104. doi: 10.1007/s10096-021-04257-8. Epub 2021 Apr 24. Eur J Clin Microbiol Infect Dis. 2021. PMID: 33895886
-
Composition and diversity analysis of the lung microbiome in patients with suspected ventilator-associated pneumonia.Crit Care. 2022 Jul 6;26(1):203. doi: 10.1186/s13054-022-04068-z. Crit Care. 2022. PMID: 35794610 Free PMC article.
-
Rapid analysis of bacterial composition in prosthetic joint infection by 16S rRNA metagenomic sequencing.Bone Joint Res. 2019 Sep 3;8(8):367-377. doi: 10.1302/2046-3758.88.BJR-2019-0003.R2. eCollection 2019 Aug. Bone Joint Res. 2019. PMID: 31537994 Free PMC article.
-
Identification of bacterial pathogens in sudden unexpected death in infancy and childhood using 16S rRNA gene sequencing.Front Microbiol. 2023 Jun 15;14:1171670. doi: 10.3389/fmicb.2023.1171670. eCollection 2023. Front Microbiol. 2023. PMID: 37396359 Free PMC article.
References
-
- Cook D. Ventilator associated pneumonia: perspectives on the burden of illness. Intensive Care Med. 2000;26(Suppl 1):S31–7. - PubMed
-
- Heyland DK, Novak F, Drover JW, Jain M, Su X, Suchner U. Should immunonutrition become routine in critically ill patients? A systematic review of the evidence. JAMA. 2001;286:944–953. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
