Identification of Distinct Breast Cancer Stem Cell Populations Based on Single-Cell Analyses of Functionally Enriched Stem and Progenitor Pools
- PMID: 26771357
- PMCID: PMC4719187
- DOI: 10.1016/j.stemcr.2015.12.006
Identification of Distinct Breast Cancer Stem Cell Populations Based on Single-Cell Analyses of Functionally Enriched Stem and Progenitor Pools
Abstract
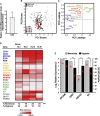
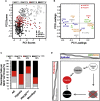
The identification of breast cancer cell subpopulations featuring truly malignant stem cell qualities is a challenge due to the complexity of the disease and lack of general markers. By combining extensive single-cell gene expression profiling with three functional strategies for cancer stem cell enrichment including anchorage-independent culture, hypoxia, and analyses of low-proliferative, label-retaining cells derived from mammospheres, we identified distinct stem cell clusters in breast cancer. Estrogen receptor (ER)α+ tumors featured a clear hierarchical organization with switch-like and gradual transitions between different clusters, illustrating how breast cancer cells transfer between discrete differentiation states in a sequential manner. ERα- breast cancer showed less prominent clustering but shared a quiescent cancer stem cell pool with ERα+ cancer. The cellular organization model was supported by single-cell data from primary tumors. The findings allow us to understand the organization of breast cancers at the single-cell level, thereby permitting better identification and targeting of cancer stem cells.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Badve S., Nakshatri H. Breast-cancer stem cells—beyond semantics. Lancet Oncol. 2012;13:e43–48. - PubMed
-
- Charafe-Jauffret E., Ginestier C., Monville F., Finetti P., Adelaide J., Cervera N., Fekairi S., Xerri L., Jacquemier J., Birnbaum D. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

