Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance
- PMID: 26771497
- PMCID: PMC4724865
- DOI: 10.1016/j.cell.2015.11.062
Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance
Abstract
Large-scale genomic studies have identified multiple somatic aberrations in breast cancer, including copy number alterations and point mutations. Still, identifying causal variants and emergent vulnerabilities that arise as a consequence of genetic alterations remain major challenges. We performed whole-genome small hairpin RNA (shRNA) "dropout screens" on 77 breast cancer cell lines. Using a hierarchical linear regression algorithm to score our screen results and integrate them with accompanying detailed genetic and proteomic information, we identify vulnerabilities in breast cancer, including candidate "drivers," and reveal general functional genomic properties of cancer cells. Comparisons of gene essentiality with drug sensitivity data suggest potential resistance mechanisms, effects of existing anti-cancer drugs, and opportunities for combination therapy. Finally, we demonstrate the utility of this large dataset by identifying BRD4 as a potential target in luminal breast cancer and PIK3CA mutations as a resistance determinant for BET-inhibitors.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
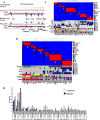


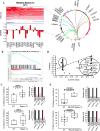


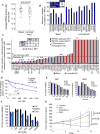
References
-
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

