A Proteome-wide Fission Yeast Interactome Reveals Network Evolution Principles from Yeasts to Human
- PMID: 26771498
- PMCID: PMC4715267
- DOI: 10.1016/j.cell.2015.11.037
A Proteome-wide Fission Yeast Interactome Reveals Network Evolution Principles from Yeasts to Human
Abstract
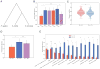
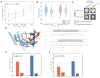
Here, we present FissionNet, a proteome-wide binary protein interactome for S. pombe, comprising 2,278 high-quality interactions, of which ∼ 50% were previously not reported in any species. FissionNet unravels previously unreported interactions implicated in processes such as gene silencing and pre-mRNA splicing. We developed a rigorous network comparison framework that accounts for assay sensitivity and specificity, revealing extensive species-specific network rewiring between fission yeast, budding yeast, and human. Surprisingly, although genes are better conserved between the yeasts, S. pombe interactions are significantly better conserved in human than in S. cerevisiae. Our framework also reveals that different modes of gene duplication influence the extent to which paralogous proteins are functionally repurposed. Finally, cross-species interactome mapping demonstrates that coevolution of interacting proteins is remarkably prevalent, a result with important implications for studying human disease in model organisms. Overall, FissionNet is a valuable resource for understanding protein functions and their evolution.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures








References
-
- Arellano M, Coll PM, Yang W, Duran A, Tamanoi F, Perez P. Characterization of the geranylgeranyl transferase type I from Schizosaccharomyces pombe. Mol Microbiol. 1998;29:1357–1367. - PubMed
-
- Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat Biotechnol. 2004;22:78–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG001715/HG/NHGRI NIH HHS/United States
- GM098634/GM/NIGMS NIH HHS/United States
- GM104424/GM/NIGMS NIH HHS/United States
- U01 HG001715/HG/NHGRI NIH HHS/United States
- P50 HG004233/HG/NHGRI NIH HHS/United States
- R01 GM098634/GM/NIGMS NIH HHS/United States
- U41 HG001715/HG/NHGRI NIH HHS/United States
- HG004233/HG/NHGRI NIH HHS/United States
- R01 GM105668/GM/NIGMS NIH HHS/United States
- R01 GM097358/GM/NIGMS NIH HHS/United States
- 13586/CRUK_/Cancer Research UK/United Kingdom
- GM097358/GM/NIGMS NIH HHS/United States
- 09-0658/AICR_/Worldwide Cancer Research/United Kingdom
- R01 GM104424/GM/NIGMS NIH HHS/United States
- HG001715/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

