Unfolding the HIV-1 reverse transcriptase RNase H domain--how to lose a molecular tug-of-war
- PMID: 26773054
- PMCID: PMC4770237
- DOI: 10.1093/nar/gkv1538
Unfolding the HIV-1 reverse transcriptase RNase H domain--how to lose a molecular tug-of-war
Abstract
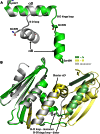
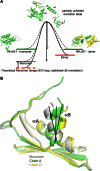
Formation of the mature HIV-1 reverse transcriptase (RT) p66/p51 heterodimer requires subunit-specific processing of the p66/p66' homodimer precursor. Since the ribonuclease H (RH) domain contains an occult cleavage site located near its center, cleavage must occur either prior to folding or subsequent to unfolding. Recent NMR studies have identified a slow, subunit-specific RH domain unfolding process proposed to result from a residue tug-of-war between the polymerase and RH domains on the functionally inactive, p66' subunit. Here, we describe a structural comparison of the isolated RH domain with a domain swapped RH dimer that reveals several intrinsically destabilizing characteristics of the isolated domain that facilitate excursions of Tyr427 from its binding pocket and separation of helices B and D. These studies provide independent support for the subunit-selective RH domain unfolding pathway in which instability of the Tyr427 binding pocket facilitates its release followed by domain transfer, acting as a trigger for further RH domain destabilization and subsequent unfolding. As further support for this pathway, NMR studies demonstrate that addition of an RH active site-directed isoquinolone ligand retards the subunit-selective RH' domain unfolding behavior of the p66/p66' homodimer. This study demonstrates the feasibility of directly targeting RT maturation with therapeutics.
Published by Oxford University Press on behalf of Nucleic Acids Research 2016. This work is written by US Government employees and is in the public domain in the US.
Figures

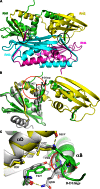



Similar articles
-
HIV-1 Reverse Transcriptase: A Metamorphic Protein with Three Stable States.Structure. 2019 Mar 5;27(3):420-426. doi: 10.1016/j.str.2018.11.011. Epub 2019 Jan 10. Structure. 2019. PMID: 30639227 Free PMC article. Review.
-
Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.Nucleic Acids Res. 2014 Apr;42(8):5361-77. doi: 10.1093/nar/gku143. Epub 2014 Feb 25. Nucleic Acids Res. 2014. PMID: 24574528 Free PMC article.
-
Asymmetric conformational maturation of HIV-1 reverse transcriptase.Elife. 2015 Jun 3;4:e06359. doi: 10.7554/eLife.06359. Elife. 2015. PMID: 26037594 Free PMC article.
-
Human immunodeficiency virus type 1 reverse transcriptase dimer destabilization by 1-[Spiro[4"-amino-2",2" -dioxo-1",2" -oxathiole-5",3'-[2', 5'-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]]]-3-ethylthy mine.Biochemistry. 2000 Feb 15;39(6):1427-33. doi: 10.1021/bi991682+. Biochemistry. 2000. PMID: 10684624
-
Structural Maturation of HIV-1 Reverse Transcriptase-A Metamorphic Solution to Genomic Instability.Viruses. 2016 Sep 27;8(10):260. doi: 10.3390/v8100260. Viruses. 2016. PMID: 27690082 Free PMC article. Review.
Cited by
-
HIV-1 Reverse Transcriptase: A Metamorphic Protein with Three Stable States.Structure. 2019 Mar 5;27(3):420-426. doi: 10.1016/j.str.2018.11.011. Epub 2019 Jan 10. Structure. 2019. PMID: 30639227 Free PMC article. Review.
-
Evolving understanding of HIV-1 reverse transcriptase structure, function, inhibition, and resistance.Curr Opin Struct Biol. 2020 Apr;61:113-123. doi: 10.1016/j.sbi.2019.11.011. Epub 2020 Jan 11. Curr Opin Struct Biol. 2020. PMID: 31935541 Free PMC article. Review.
-
Identification of drivers for the metamorphic transition of HIV-1 reverse transcriptase.Biochem J. 2017 Sep 24;474(19):3321-3338. doi: 10.1042/BCJ20170480. Biochem J. 2017. PMID: 28811321 Free PMC article.
-
Determinants of Active-Site Inhibitor Interaction with HIV-1 RNase H.ACS Infect Dis. 2019 Nov 8;5(11):1963-1974. doi: 10.1021/acsinfecdis.9b00300. Epub 2019 Oct 2. ACS Infect Dis. 2019. PMID: 31577424 Free PMC article.
-
Conformational Changes in HIV-1 Reverse Transcriptase that Facilitate Its Maturation.Structure. 2019 Oct 1;27(10):1581-1593.e3. doi: 10.1016/j.str.2019.08.004. Epub 2019 Aug 27. Structure. 2019. PMID: 31471129 Free PMC article.
References
-
- Jochmans D. NovelHIV-1 reverse transcriptase inhibitors. Virus Res. 2008;134:171–185. - PubMed
-
- Ibe S., Sugiura W. Clinical significance of HIV reverse-transcriptase inhibitor-resistance mutations. Future Microbiol. 2011;6:295–315. - PubMed
-
- Menendez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 2010;85:210–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

