CrossHub: a tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms
- PMID: 26773058
- PMCID: PMC4838350
- DOI: 10.1093/nar/gkv1478
CrossHub: a tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms
Abstract
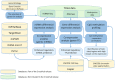
The contribution of different mechanisms to the regulation of gene expression varies for different tissues and tumors. Complementation of predicted mRNA-miRNA and gene-transcription factor (TF) relationships with the results of expression correlation analyses derived for specific tumor types outlines the interactions with functional impact in the current biomaterial. We developed CrossHub software, which enables two-way identification of most possible TF-gene interactions: on the basis of ENCODE ChIP-Seq binding evidence or Jaspar prediction and co-expression according to the data of The Cancer Genome Atlas (TCGA) project, the largest cancer omics resource. Similarly, CrossHub identifies mRNA-miRNA pairs with predicted or validated binding sites (TargetScan, mirSVR, PicTar, DIANA microT, miRTarBase) and strong negative expression correlations. We observed partial consistency between ChIP-Seq or miRNA target predictions and gene-TF/miRNA co-expression, demonstrating a link between these indicators. Additionally, CrossHub expression-methylation correlation analysis can be used to identify hypermethylated CpG sites or regions with the greatest potential impact on gene expression. Thus, CrossHub is capable of outlining molecular portraits of a specific gene and determining the three most common sources of expression regulation: promoter/enhancer methylation, miRNA interference and TF-mediated activation or repression. CrossHub generates formatted Excel workbooks with the detailed results. CrossHub is freely available athttps://sourceforge.net/projects/crosshub/.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
[Overexpression of microRNAs miR-9, -98, and -199 Correlates with the Downregulation of HK2 Expression in Colorectal Cancer].Mol Biol (Mosk). 2018 Mar-Apr;52(2):220-230. doi: 10.7868/S0026898418020052. Mol Biol (Mosk). 2018. PMID: 29695690 Russian.
-
DIANA-mirExTra v2.0: Uncovering microRNAs and transcription factors with crucial roles in NGS expression data.Nucleic Acids Res. 2016 Jul 8;44(W1):W128-34. doi: 10.1093/nar/gkw455. Epub 2016 May 20. Nucleic Acids Res. 2016. PMID: 27207881 Free PMC article.
-
Bioinformatics method to predict two regulation mechanism: TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer.Cell Biochem Biophys. 2014 Dec;70(3):1849-58. doi: 10.1007/s12013-014-0142-y. Cell Biochem Biophys. 2014. PMID: 25087086
-
"Promoter array" studies identify cohorts of genes directly regulated by methylation, copy number change, or transcription factor binding in human cancer cells.Ann N Y Acad Sci. 2005 Nov;1058:162-85. doi: 10.1196/annals.1359.024. Ann N Y Acad Sci. 2005. PMID: 16394135 Review.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Characterization and functional analyses of the human HTR1A gene: 5' regulatory region modulates gene expression in vitro.BMC Genet. 2018 Dec 29;19(1):115. doi: 10.1186/s12863-018-0708-6. BMC Genet. 2018. PMID: 30594152 Free PMC article.
-
Differential expression of alternatively spliced transcripts related to energy metabolism in colorectal cancer.BMC Genomics. 2016 Dec 28;17(Suppl 14):1011. doi: 10.1186/s12864-016-3351-5. BMC Genomics. 2016. PMID: 28105922 Free PMC article.
-
The CIMP-high phenotype is associated with energy metabolism alterations in colon adenocarcinoma.BMC Med Genet. 2019 Apr 9;20(Suppl 1):52. doi: 10.1186/s12881-019-0771-5. BMC Med Genet. 2019. PMID: 30967137 Free PMC article.
-
Tumor suppressor properties of the small C-terminal domain phosphatases in non-small cell lung cancer.Biosci Rep. 2019 Dec 20;39(12):BSR20193094. doi: 10.1042/BSR20193094. Biosci Rep. 2019. PMID: 31774910 Free PMC article.
-
Deep Sequencing Revealed a CpG Methylation Pattern Associated With ALDH1L1 Suppression in Breast Cancer.Front Genet. 2018 May 15;9:169. doi: 10.3389/fgene.2018.00169. eCollection 2018. Front Genet. 2018. PMID: 29868117 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

