A TonB-Dependent Transporter Is Responsible for Methanobactin Uptake by Methylosinus trichosporium OB3b
- PMID: 26773085
- PMCID: PMC4784032
- DOI: 10.1128/AEM.03884-15
A TonB-Dependent Transporter Is Responsible for Methanobactin Uptake by Methylosinus trichosporium OB3b
Abstract
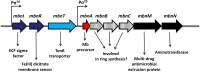
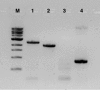
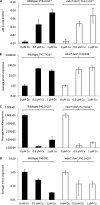
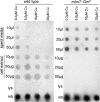
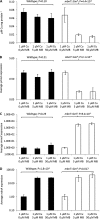
Methanobactin, a small modified polypeptide synthesized by methanotrophs for copper uptake, has been found to be chromosomally encoded. The gene encoding the polypeptide precursor of methanobactin, mbnA, is part of a gene cluster that also includes several genes encoding proteins of unknown function (but speculated to be involved in methanobactin formation) as well as mbnT, which encodes a TonB-dependent transporter hypothesized to be responsible for methanobactin uptake. To determine if mbnT is truly responsible for methanobactin uptake, a knockout was constructed in Methylosinus trichosporium OB3b using marker exchange mutagenesis. The resulting M. trichosporium mbnT::Gm(r) mutant was found to be able to produce methanobactin but was unable to internalize it. Further, if this mutant was grown in the presence of copper and exogenous methanobactin, copper uptake was significantly reduced. Expression of mmoX and pmoA, encoding polypeptides of the soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO), respectively, also changed significantly when methanobactin was added, which indicates that the mutant was unable to collect copper under these conditions. Copper uptake and gene expression, however, were not affected in wild-type M. trichosporium OB3b, indicating that the TonB-dependent transporter encoded by mbnT is responsible for methanobactin uptake and that methanobactin is a key mechanism used by methanotrophs for copper uptake. When the mbnT::Gm(r) mutant was grown under a range of copper concentrations in the absence of methanobactin, however, the phenotype of the mutant was indistinguishable from that of wild-type M. trichosporium OB3b, indicating that this methanotroph has multiple mechanisms for copper uptake.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Myhre G, Shindell D, Breon F-M, Collins W, Fuglestvedt J, Huang J, Koch D, Lamarque JF, Lee D, Mendoza B, Nakajima T, Robock A, Stephens G, Takamura T, Zhang H. 2013. Anthropogenic and natural radiative forcing, p 659–740. In Stocker TF, Qin D, Plattner GK, Tignor MMB, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM (ed), Climate change 2013: the physical science basis. Cambridge University Press, Cambridge, United Kingdom.
-
- Chowdury TR, Dick RP. 2013. Ecology of aerobic methanotrophs in controlling methane fluxes from wetlands. Appl Soil Ecol 65:8–22. doi: 10.1016/j.apsoil.2012.12.014. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

