Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α
- PMID: 26774474
- PMCID: PMC4731243
- DOI: 10.1016/j.celrep.2015.12.043
Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α
Abstract
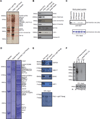
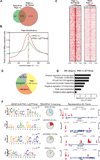
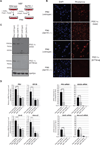
The Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α) is a transcriptional co-activator that plays a central role in adapted metabolic responses. PGC-1α is dynamically methylated and unmethylated at the residue K779 by the methyltransferase SET7/9 and the Lysine Specific Demethylase 1A (LSD1), respectively. Interactions of methylated PGC-1α[K779me] with the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex, the Mediator members MED1 and MED17, and the NOP2/Sun RNA methytransferase 7 (NSUN7) reinforce transcription, and are concomitant with the m(5)C mark on enhancer RNAs (eRNAs). Consistently, loss of Set7/9 and NSun7 in liver cell model systems resulted in depletion of the PGC-1α target genes Pfkl, Sirt5, Idh3b, and Hmox2, which was accompanied by a decrease in the eRNAs levels associated with these loci. Enrichment of m(5)C within eRNA species coincides with metabolic stress of fasting in vivo. Collectively, these findings illustrate the complex epigenetic circuitry imposed by PGC-1α at the eRNA level to fine-tune energy metabolism.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Aubert B, Karyotakis Y, Lees JP, Poireau V, Prencipe E, Prudent X, Tisserand V, Garra Tico J, Grauges E, Martinelli M, et al. Search for dimuon decays of a light scalar boson in radiative transitions Upsilon-->gammaA0. Phys Rev Lett. 2009;103:081803. - PubMed
-
- Babitzke P. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol. 2004;7:132–139. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA154903/CA/NCI NIH HHS/United States
- R01 HL103967/HL/NHLBI NIH HHS/United States
- DK084434/DK/NIDDK NIH HHS/United States
- R01 DK084434/DK/NIDDK NIH HHS/United States
- R01 HL067099/HL/NHLBI NIH HHS/United States
- HL067099/HL/NHLBI NIH HHS/United States
- R01 GM119189/GM/NIGMS NIH HHS/United States
- RC1 DA028776/DA/NIDA NIH HHS/United States
- R01 GM089778/GM/NIGMS NIH HHS/United States
- R01 AI124465/AI/NIAID NIH HHS/United States
- R01 CA087658/CA/NCI NIH HHS/United States
- R01 CA154809/CA/NCI NIH HHS/United States
- HL103967/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

