The design and analysis of transposon insertion sequencing experiments
- PMID: 26775926
- PMCID: PMC5099075
- DOI: 10.1038/nrmicro.2015.7
The design and analysis of transposon insertion sequencing experiments
Abstract
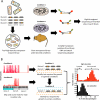
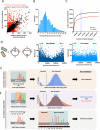
Transposon insertion sequencing (TIS) is a powerful approach that can be extensively applied to the genome-wide definition of loci that are required for bacterial growth under diverse conditions. However, experimental design choices and stochastic biological processes can heavily influence the results of TIS experiments and affect downstream statistical analysis. In this Opinion article, we discuss TIS experimental parameters and how these factors relate to the benefits and limitations of the various statistical frameworks that can be applied to the computational analysis of TIS data.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

