Enhanced MAPK signaling drives ETS1-mediated induction of miR-29b leading to downregulation of TET1 and changes in epigenetic modifications in a subset of lung SCC
- PMID: 26776158
- PMCID: PMC4994018
- DOI: 10.1038/onc.2015.499
Enhanced MAPK signaling drives ETS1-mediated induction of miR-29b leading to downregulation of TET1 and changes in epigenetic modifications in a subset of lung SCC
Abstract
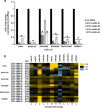
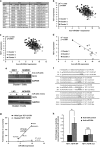
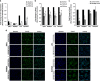
Non-small-cell lung cancer is the leading cause of cancer death worldwide and is comprised of several histological subtypes, the two most common being adenocarcinoma (AC) and squamous cell carcinoma (SCC). Targeted therapies have successfully improved response rates in patients with AC tumors. However, the majority of SCC tumors lack specific targetable mutations, making development of new treatment paradigms for this disease challenging. In the present study, we used iterative non-negative matrix factorization, an unbiased clustering method, on mRNA expression data from the cancer genome atlas (TCGA) and a panel of 24 SCC cell lines to classify three disease segments within SCC. Analysis of gene set enrichment and drug sensitivity identified an immune-evasion subtype that showed increased sensitivity to nuclear factor-κB and mitogen-activated protein kinase (MAPK) inhibition, a replication-stress associated subtype that showed increased sensitivity to ataxia telangiectasia inhibition, and a neuroendocrine-associated subtype that showed increased sensitivity to phosphoinositide 3-kinase and fibroblast growth factor receptor inhibition. Additionally, each of these subtypes exhibited a unique microRNA expression profile. Focusing on the immune-evasion subtype, bioinformatic analysis of microRNA promoters revealed enrichment for binding sites for the MAPK-driven ETS1 transcription factor. Indeed, we found that knockdown of ETS1 led to upregulation of eight microRNAs and downregulation of miR-29b in the immune-evasion subtype. Mechanistically, we found that miR-29b targets the DNA-demethylating enzyme, TET1, for downregulation resulting in decreased 5-hmC epigenetic modifications. Moreover, inhibition of MAPK signaling by gefitinib led to decreased ETS1 and miR-29b expression with a corresponding increase in TET1 expression and increase in 5-hmC. Collectively, our work identifies three subtypes of lung SCC that differ in drug sensitivity and shows a novel mechanism of miR-29b regulation by MAPK-driven ETS1 expression which leads to downstream changes in TET1-mediated epigenetic modifications.
Conflict of interest statement
All authors are employees of AstraZeneca.
Figures




References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. - PubMed
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531–537. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. - PubMed
-
- Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 2006; 24: 5079–5090. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

