Comparative Genomics of Two ST 195 Carbapenem-Resistant Acinetobacter baumannii with Different Susceptibility to Polymyxin Revealed Underlying Resistance Mechanism
- PMID: 26779129
- PMCID: PMC4700137
- DOI: 10.3389/fmicb.2015.01445
Comparative Genomics of Two ST 195 Carbapenem-Resistant Acinetobacter baumannii with Different Susceptibility to Polymyxin Revealed Underlying Resistance Mechanism
Abstract
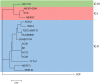
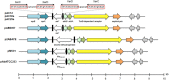
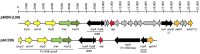
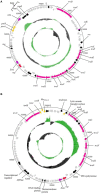
Acinetobacter baumannii is a Gram-negative nosocomial pathogen of importance due to its uncanny ability to acquire resistance to most antimicrobials. These include carbapenems, which are the drugs of choice for treating A. baumannii infections, and polymyxins, the drugs of last resort. Whole genome sequencing was performed on two clinical carbapenem-resistant A. baumannii AC29 and AC30 strains which had an indistinguishable ApaI pulsotype but different susceptibilities to polymyxin. Both genomes consisted of an approximately 3.8 Mbp circular chromosome each and several plasmids. AC29 (susceptible to polymyxin) and AC30 (resistant to polymyxin) belonged to the ST195 lineage and are phylogenetically clustered under the International Clone II (IC-II) group. An AbaR4-type resistance island (RI) interrupted the comM gene in the chromosomes of both strains and contained the bla OXA-23 carbapenemase gene and determinants for tetracycline and streptomycin resistance. AC29 harbored another copy of bla OXA-23 in a large (~74 kb) conjugative plasmid, pAC29b, but this gene was absent in a similar plasmid (pAC30c) found in AC30. A 7 kb Tn1548::armA RI which encodes determinants for aminoglycoside and macrolide resistance, is chromosomally-located in AC29 but found in a 16 kb plasmid in AC30, pAC30b. Analysis of known determinants for polymyxin resistance in AC30 showed mutations in the pmrA gene encoding the response regulator of the two-component pmrAB signal transduction system as well as in the lpxD, lpxC, and lpsB genes that encode enzymes involved in the biosynthesis of lipopolysaccharide (LPS). Experimental evidence indicated that impairment of LPS along with overexpression of pmrAB may have contributed to the development of polymyxin resistance in AC30. Cloning of a novel variant of the bla AmpC gene from AC29 and AC30, and its subsequent expression in E. coli also indicated its likely function as an extended-spectrum cephalosporinase.
Keywords: Acinetobacter baumannii; carbapenem-resistant; polymyxin; resistance island; resistance mechanisms; whole genome sequencing.
Figures


References
-
- Antunes L. C. S., Imperi F., Towner K. J., Visca P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284. 10.1016/j.resmic.2010.10.010 - DOI - PubMed
-
- Arroyo L. A., Herrera C. M., Fernandez L., Hankins J. V., Trent M. S., Hancock R. E. W. (2011). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55, 3743–3751. 10.1128/AAC.00256-11 - DOI - PMC - PubMed
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. D., Smith J. A., et al. (2002). Short Protocols in Molecular Biology (5th Edn). John Wiley & Sons.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

