New Insights into the Classification and Integration Specificity of Streptococcus Integrative Conjugative Elements through Extensive Genome Exploration
- PMID: 26779141
- PMCID: PMC4701971
- DOI: 10.3389/fmicb.2015.01483
New Insights into the Classification and Integration Specificity of Streptococcus Integrative Conjugative Elements through Extensive Genome Exploration
Abstract
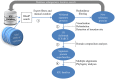
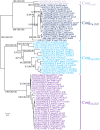
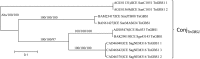
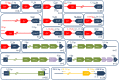
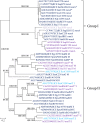
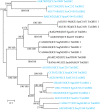
Recent genome analyses suggest that integrative and conjugative elements (ICEs) are widespread in bacterial genomes and therefore play an essential role in horizontal transfer. However, only a few of these elements are precisely characterized and correctly delineated within sequenced bacterial genomes. Even though previous analysis showed the presence of ICEs in some species of Streptococci, the global prevalence and diversity of ICEs was not analyzed in this genus. In this study, we searched for ICEs in the completely sequenced genomes of 124 strains belonging to 27 streptococcal species. These exhaustive analyses revealed 105 putative ICEs and 26 slightly decayed elements whose limits were assessed and whose insertion site was identified. These ICEs were grouped in seven distinct unrelated or distantly related families, according to their conjugation modules. Integration of these streptococcal ICEs is catalyzed either by a site-specific tyrosine integrase, a low-specificity tyrosine integrase, a site-specific single serine integrase, a triplet of site-specific serine integrases or a DDE transposase. Analysis of their integration site led to the detection of 18 target-genes for streptococcal ICE insertion including eight that had not been identified previously (ftsK, guaA, lysS, mutT, rpmG, rpsI, traG, and ebfC). It also suggests that all specificities have evolved to minimize the impact of the insertion on the host. This overall analysis of streptococcal ICEs emphasizes their prevalence and diversity and demonstrates that exchanges or acquisitions of conjugation and recombination modules are frequent.
Keywords: Streptococcus; T4SS; integrase; integration site; integrative and conjugative elements.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

