Expanding our Understanding of the Seaweed Holobiont: RNA Viruses of the Red Alga Delisea pulchra
- PMID: 26779145
- PMCID: PMC4705237
- DOI: 10.3389/fmicb.2015.01489
Expanding our Understanding of the Seaweed Holobiont: RNA Viruses of the Red Alga Delisea pulchra
Abstract
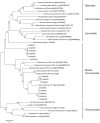
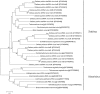
Marine seaweeds are holobionts comprised of the macroalgal hosts and their associated microbiota. While the composition of the bacterial component of seaweed microbiomes is increasingly studied, almost nothing is known about the presence, diversity and composition of viruses in macroalgae in situ. In this study, we characterize for the first time the viruses associated with a red macroalga, Delisea pulchra. Using transmission electron microscopy we identified diverse morphotypes of virus-like particles in D. pulchra ranging from icosahedral to bacilliform to coiled pleomorphic as well as bacteriophages. Virome sequencing revealed the presence of a diverse group of dsRNA viruses affiliated to the genus Totivirus, known to infect plant pathogenic fungi. We further identified a ssRNA virus belonging to the order Picornavirales with a close phylogenetic relationship to a pathogenic virus infecting marine diatoms. The results of this study shed light on a so far neglected part of the seaweed holobiont, and suggest that some of the identified viruses may be possible pathogens for a host that is already known to be significantly impacted by bacterial infections.
Keywords: alga; disease; fungi; macroalga; metaorganism; pathogen; seaweed; viromes.
Figures



Similar articles
-
Multiple opportunistic pathogens can cause a bleaching disease in the red seaweed Delisea pulchra.Environ Microbiol. 2016 Nov;18(11):3962-3975. doi: 10.1111/1462-2920.13403. Epub 2016 Jul 15. Environ Microbiol. 2016. PMID: 27337296
-
Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed Delisea pulchra.PLoS One. 2012;7(12):e50854. doi: 10.1371/journal.pone.0050854. Epub 2012 Dec 3. PLoS One. 2012. PMID: 23226544 Free PMC article.
-
A glutathione peroxidase (GpoA) plays a role in the pathogenicity of Nautella italica strain R11 towards the red alga Delisea pulchra.FEMS Microbiol Ecol. 2015 Apr;91(4):fiv021. doi: 10.1093/femsec/fiv021. Epub 2015 Feb 26. FEMS Microbiol Ecol. 2015. PMID: 25764469
-
Chemical mediation of ternary interactions between marine holobionts and their environment as exemplified by the red alga Delisea pulchra.J Chem Ecol. 2012 May;38(5):442-50. doi: 10.1007/s10886-012-0119-5. Epub 2012 Apr 25. J Chem Ecol. 2012. PMID: 22527059 Review.
-
Progress and future directions for seaweed holobiont research.New Phytol. 2024 Oct;244(2):364-376. doi: 10.1111/nph.20018. Epub 2024 Aug 13. New Phytol. 2024. PMID: 39137959 Review.
Cited by
-
The RNA virosphere: How big and diverse is it?Environ Microbiol. 2023 Jan;25(1):209-215. doi: 10.1111/1462-2920.16312. Epub 2022 Dec 28. Environ Microbiol. 2023. PMID: 36511833 Free PMC article. No abstract available.
-
Diverse RNA Viruses Associated with Diatom, Eustigmatophyte, Dinoflagellate, and Rhodophyte Microalgae Cultures.J Virol. 2022 Oct 26;96(20):e0078322. doi: 10.1128/jvi.00783-22. Epub 2022 Oct 3. J Virol. 2022. PMID: 36190242 Free PMC article.
-
The seaweed holobiont: from microecology to biotechnological applications.Microb Biotechnol. 2022 Mar;15(3):738-754. doi: 10.1111/1751-7915.14014. Epub 2022 Feb 8. Microb Biotechnol. 2022. PMID: 35137526 Free PMC article. Review.
-
Virome Variation during Sea Star Wasting Disease Progression in Pisaster ochraceus (Asteroidea, Echinodermata).Viruses. 2020 Nov 20;12(11):1332. doi: 10.3390/v12111332. Viruses. 2020. PMID: 33233680 Free PMC article.
-
High-Quality Resolution of the Outbreak-Related Zika Virus Genome and Discovery of New Viruses Using Ion Torrent-Based Metatranscriptomics.Viruses. 2020 Jul 21;12(7):782. doi: 10.3390/v12070782. Viruses. 2020. PMID: 32708079 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

