Expanding the World of Marine Bacterial and Archaeal Clades
- PMID: 26779174
- PMCID: PMC4705458
- DOI: 10.3389/fmicb.2015.01524
Expanding the World of Marine Bacterial and Archaeal Clades
Abstract
Determining which microbial taxa are out there, where they live, and what they are doing is a driving approach in marine microbial ecology. The importance of these questions is underlined by concerted, large-scale, and global ocean sampling initiatives, for example the International Census of Marine Microbes, Ocean Sampling Day, or Tara Oceans. Given decades of effort, we know that the large majority of marine Bacteria and Archaea belong to about a dozen phyla. In addition to the classically culturable Bacteria and Archaea, at least 50 "clades," at different taxonomic depths, exist. These account for the majority of marine microbial diversity, but there is still an underexplored and less abundant portion remaining. We refer to these hitherto unrecognized clades as unknown, as their boundaries, names, and classifications are not available. In this work, we were able to characterize up to 92 of these unknown clades found within the bacterial and archaeal phylogenetic diversity currently reported for marine water column environments. We mined the SILVA 16S rRNA gene datasets for sequences originating from the marine water column. Instead of the usual subjective taxa delineation and nomenclature methods, we applied the candidate taxonomic unit (CTU) circumscription system, along with a standardized nomenclature to the sequences in newly constructed phylogenetic trees. With this new phylogenetic and taxonomic framework, we performed an analysis of ICoMM rRNA gene amplicon datasets to gain insights into the global distribution of the new marine clades, their ecology, biogeography, and interaction with oceanographic variables. Most of the new clades we identified were interspersed by known taxa with cultivated members, whose genome sequences are available. This result encouraged us to perform metabolic predictions for the novel marine clades using the PICRUSt approach. Our work also provides an update on the taxonomy of several phyla and widely known marine clades as our CTU approach breaks down these randomly lumped clades into smaller objectively calculated subgroups. Finally, all taxa were classified and named following standards compatible with the Bacteriological Code rules, enhancing their digitization, and comparability with future microbial ecological and taxonomy studies.
Keywords: bacterial phylogeny; bacterial taxonomy; bacterioplankton; ecology; marine; rare taxa.
Figures
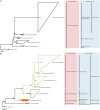
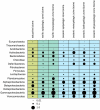



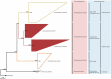
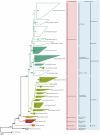
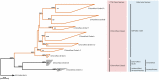

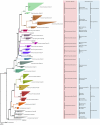
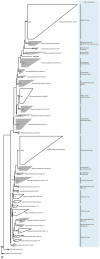

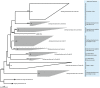
References
-
- Alain K., Olagnon M., Desbruyères D., Pagé A., Barbier G., Juniper S. K., et al. . (2002). Phylogenetic characterization of the bacterial assemblage associated with mucous secretions of the hydrothermal vent polychaete Paralvinella palmiformis. FEMS Microbiol. Ecol. 42, 463–476. 10.1111/j.1574-6941.2002.tb01035.x - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

