Cross-Talk between AURKA and Plk1 in Mitotic Entry and Spindle Assembly
- PMID: 26779436
- PMCID: PMC4688340
- DOI: 10.3389/fonc.2015.00283
Cross-Talk between AURKA and Plk1 in Mitotic Entry and Spindle Assembly
Abstract
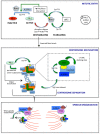
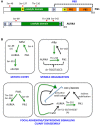
The Aurora kinase A (AURKA) is involved in different aspects of mitotic control, from mitotic entry to cytokinesis. Consistent with its pleiotropic roles, several AURKA interactors are able to modulate its activity, the best characterized being the microtubule-binding protein TPX2, the centrosomal protein Cep192, and Bora. Bora has been described as an essential cofactor of AURKA for phosphorylation-mediated activation of the mitotic kinase polo-like kinase 1 (Plk1) at the G2/M transition. A complex AURKA/Plk1 signaling axis is emerging, with multiple involved actors; recent data suggest that this control network is not restricted to mitotic entry only, but operates throughout mitosis. Here, we integrate available data from the literature to depict the complex interplay between AURKA and Plk1 in G2 and mitosis and how it contributes to their mitotic functions. We will particularly focus on how the activity of specifically localized AURKA/Plk1 pools is modulated in time and space by their reciprocal regulation to ensure the timely and coordinated unfolding of downstream mitotic events.
Keywords: G2/M transition; centrosomes; kinases; mitosis; spindle.
Figures
References
-
- Sunkel CE, Glover DM. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci (1998) 89:25–38. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

