Chlamydia Infection Across Host Species Boundaries Promotes Distinct Sets of Transcribed Anti-Apoptotic Factors
- PMID: 26779446
- PMCID: PMC4688367
- DOI: 10.3389/fcimb.2015.00096
Chlamydia Infection Across Host Species Boundaries Promotes Distinct Sets of Transcribed Anti-Apoptotic Factors
Abstract
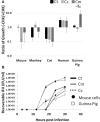
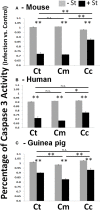
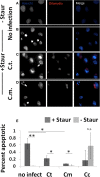
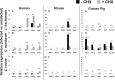
Chlamydiae, obligate intracellular bacteria, cause significant human and veterinary associated diseases. Having emerged an estimated 700-million years ago, these bacteria have twice adapted to humans as a host species, causing sexually transmitted infection (C. trachomatis) and respiratory associated disease (C. pneumoniae). The principle mechanism of host cell defense against these intracellular bacteria is the induction of cell death via apoptosis. However, in the "arms race" of co-evolution, Chlamydiae have developed mechanisms to promote cell viability and inhibit cell death. Herein we examine the impact of Chlamydiae infection across multiple host species on transcription of anti-apoptotic genes. We found mostly distinct patterns of gene expression (Mcl1 and cIAPs) elicited by each pathogen-host pair indicating Chlamydiae infection across host species boundaries does not induce a universally shared host response. Understanding species specific host-pathogen interactions is paramount to deciphering how potential pathogens become emerging diseases.
Keywords: Chlamydia; apoptosis; emerging disease; evolution; transcription.
Figures
References
-
- Allan I., Pearce J. H. (1983b). Differential amino acid utilization by Chlamydia psittaci (strain guinea pig inclusion conjunctivitis) and its regulatory effect on chlamydial growth. J. Gen. Microbiol. 129, 1991–2000. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

