Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation
- PMID: 26779602
- PMCID: PMC5790184
- DOI: 10.1038/ni.3349
Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation
Abstract
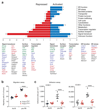
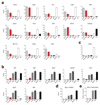
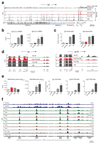
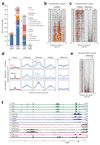
The transcription factor Blimp-1 is necessary for the generation of plasma cells. Here we studied its functions in plasmablast differentiation by identifying regulated Blimp-1 target genes. Blimp-1 promoted the migration and adhesion of plasmablasts. It directly repressed genes encoding several transcription factors and Aicda (which encodes the cytidine deaminase AID) and thus silenced B cell-specific gene expression, antigen presentation and class-switch recombination in plasmablasts. It directly activated genes, which led to increased expression of the plasma cell regulator IRF4 and proteins involved in immunoglobulin secretion. Blimp-1 induced the transcription of immunoglobulin genes by controlling the 3' enhancers of the loci encoding the immunoglobulin heavy chain (Igh) and κ-light chain (Igk) and, furthermore, regulated the post-transcriptional expression switch from the membrane-bound form of the immunoglobulin heavy chain to its secreted form by activating Ell2 (which encodes the transcription-elongation factor ELL2). Notably, Blimp-1 recruited chromatin-remodeling and histone-modifying complexes to regulate its target genes. Hence, many essential functions of plasma cells are under the control of Blimp-1.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. - PubMed
-
- Shi W, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16:663–673. - PubMed
-
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in reponse to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. - PubMed
-
- Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. - PubMed
-
- Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. - PubMed
Online Methods References
-
- Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. - PubMed
-
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. - PubMed
-
- Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. - PubMed
-
- Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

