An informatic framework for decoding protein complexes by top-down mass spectrometry
- PMID: 26780093
- PMCID: PMC4767540
- DOI: 10.1038/nmeth.3731
An informatic framework for decoding protein complexes by top-down mass spectrometry
Abstract
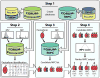
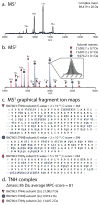
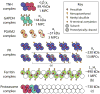
Efforts to map the human protein interactome have resulted in information about thousands of multi-protein assemblies housed in public repositories, but the molecular characterization and stoichiometry of their protein subunits remains largely unknown. Here, we report a computational search strategy that supports hierarchical top-down analysis for precise identification and scoring of multi-proteoform complexes by native mass spectrometry.
Conflict of interest statement
ProSightPC software is a commercial product for which R.T.F., R.D.L., and N.L.K. receive proceeds. M.E.B., S.R.H., and A.A.M. are employees of Thermo Fischer Scientific, the manufacturer of Q-Exactive instruments.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

