A topological and conformational stability alphabet for multipass membrane proteins
- PMID: 26780406
- PMCID: PMC6495056
- DOI: 10.1038/nchembio.2001
A topological and conformational stability alphabet for multipass membrane proteins
Abstract
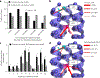
Multipass membrane proteins perform critical signal transduction and transport across membranes. How transmembrane helix (TMH) sequences encode the topology and conformational flexibility regulating these functions remains poorly understood. Here we describe a comprehensive analysis of the sequence-structure relationships at multiple interacting TMHs from all membrane proteins with structures in the Protein Data Bank (PDB). We found that membrane proteins can be deconstructed in interacting TMH trimer units, which mostly fold into six distinct structural classes of topologies and conformations. Each class is enriched in recurrent sequence motifs from functionally unrelated proteins, revealing unforeseen consensus and evolutionary conserved networks of stabilizing interhelical contacts. Interacting TMHs' topology and local protein conformational flexibility were remarkably well predicted in a blinded fashion from the identified binding-hotspot motifs. Our results reveal universal sequence-structure principles governing the complex anatomy and plasticity of multipass membrane proteins that may guide de novo structure prediction, design, and studies of folding and dynamics.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures


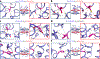

References
-
- von Heijne G Membrane-protein topology. Nat. Rev. Mol. Cell Biol 7, 909–918 (2006). - PubMed
-
- Matthews EE, Zoonens M & Engelman DM Dynamic helix interactions in transmembrane signaling. Cell 127, 447–450 (2006). - PubMed
-
- Rakoczy EP, Kiel C, McKeone R, Stricher F & Serrano L Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J. Mol. Biol 405, 584–606 (2011). - PubMed
-
- Partridge AW, Therien AG & Deber CM Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins 54, 648–656 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

