Microglial brain region-dependent diversity and selective regional sensitivities to aging
- PMID: 26780511
- PMCID: PMC4768346
- DOI: 10.1038/nn.4222
Microglial brain region-dependent diversity and selective regional sensitivities to aging
Abstract
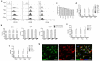
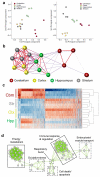
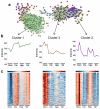
Microglia have critical roles in neural development, homeostasis and neuroinflammation and are increasingly implicated in age-related neurological dysfunction. Neurodegeneration often occurs in disease-specific, spatially restricted patterns, the origins of which are unknown. We performed to our knowledge the first genome-wide analysis of microglia from discrete brain regions across the adult lifespan of the mouse, and found that microglia have distinct region-dependent transcriptional identities and age in a regionally variable manner. In the young adult brain, differences in bioenergetic and immunoregulatory pathways were the major sources of heterogeneity and suggested that cerebellar and hippocampal microglia exist in a more immune-vigilant state. Immune function correlated with regional transcriptional patterns. Augmentation of the distinct cerebellar immunophenotype and a contrasting loss in distinction of the hippocampal phenotype among forebrain regions were key features during aging. Microglial diversity may enable regionally localized homeostatic functions but could also underlie region-specific sensitivities to microglial dysregulation and involvement in age-related neurodegeneration.
Figures





References
-
- Prinz M, Mildner A. Microglia in the CNS: immigrants from another world. Glia. 2011;59:177–187. - PubMed
-
- Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. - PubMed
-
- Paolicelli RC, et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science (New York, N.Y. 2011;333:1456–1458. - PubMed
-
- Coull JAM, et al. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. - PubMed
Publication types
MeSH terms
Grants and funding
- BBS/E/D/05191131/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L003384/1/MRC_/Medical Research Council/United Kingdom
- BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004332/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900740/MRC_/Medical Research Council/United Kingdom
- BB/I001107/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20251969/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K001744/1/MRC_/Medical Research Council/United Kingdom
- BB/J004243/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

