Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries
- PMID: 26782782
- PMCID: PMC4717575
- DOI: 10.1186/s12858-016-0057-x
Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries
Abstract
Background: The use of metagenomics in enzyme discovery constitutes a powerful approach to access to genomes of unculturable community of microorganisms and isolate novel valuable biocatalysts for use in a wide range of biotechnological and pharmaceutical fields.
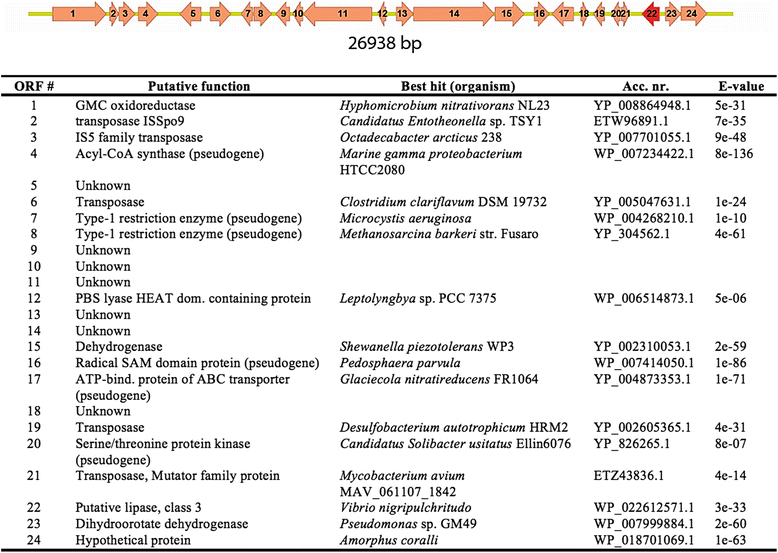
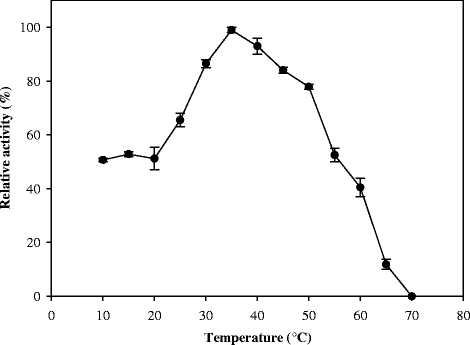
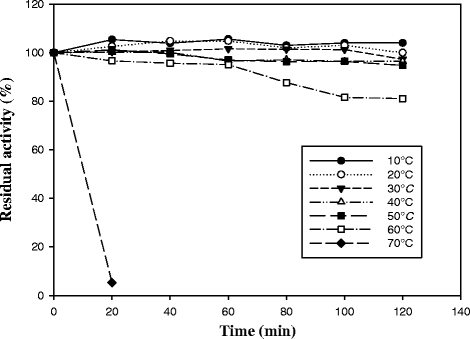
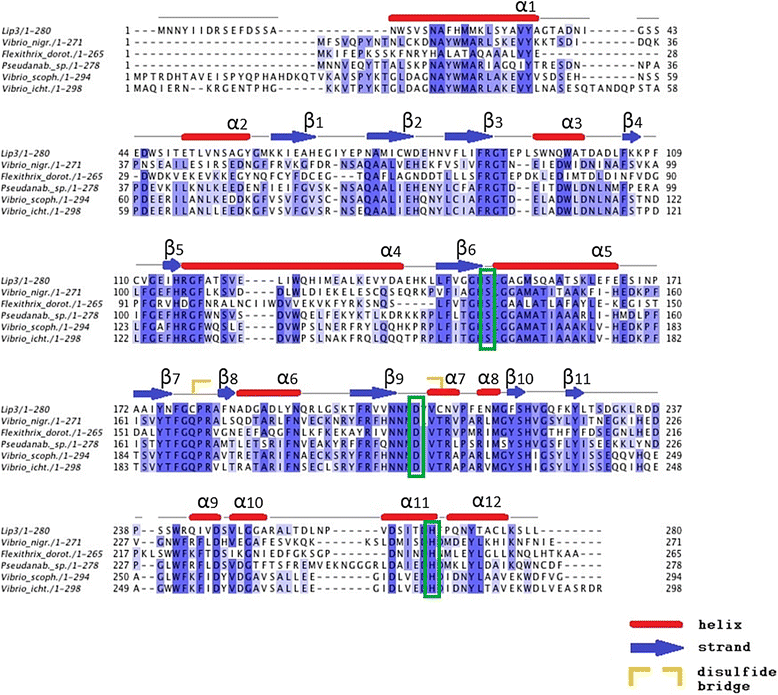
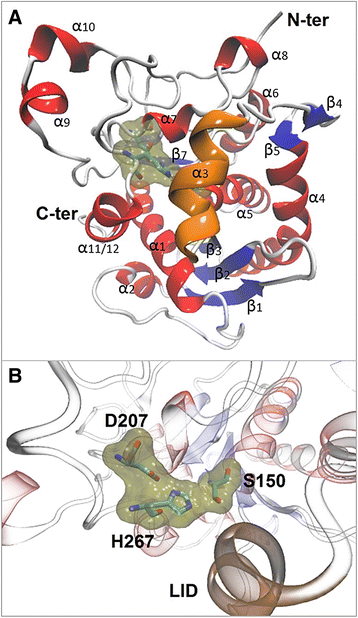
Results: Here we present a novel esterase gene (lip3) identified by functional screening of three fosmid metagenomic libraries, constructed from three marine sediment samples. The sequenced positive fosmid revealed an enzyme of 281 amino acids with similarity to class 3 lipases. The 3D modeling of Lip3 was generated by homology modeling on the basis of four lipases templates [PDB ID: 3O0D, 3NGM, 3G7N, 2QUB] to unravel structural features of this novel enzyme. The catalytic triad of Lip3 was predicted to be Asp207, His267 and the catalytic nucleophile Ser150 in a conserved pentapeptide (GXSXG). The 3D model highlighted the presence of a one-helix lid able to regulate the access of the substrate to the active site when the enzyme binds a hydrophobic interface. Moreover an analysis of the external surface of Lip3 model showed that the majority of the surface regions were hydrophobic (59.6 %) compared with homologous lipases (around 35 %) used as templates. The recombinant Lip3 esterase, expressed and purified from Escherichia coli, preferentially hydrolyzed short and medium length p-nitrophenyl esters with the best substrate being p-nitrophenyl acetate. Further characterization revealed a temperature optimum of 35 °C and a pH optimum of 8.0. Lip3 exhibits a broad temperature stability range and tolerates the presence of DTT, EDTA, PMSF, β-mercaptoethanol and high concentrations of salt. The enzyme was also highly activated by NaCl.
Conclusions: The biochemical characterization and homology model reveals a novel esterase originating from the marine Arctic metagenomics libraries with features of a cold-active, relatively thermostable and highly halotolerant enzyme. Taken together, these results suggest that this esterase could be a highly valuable candidate for biotechnological applications such as organic synthesis reactions and cheese ripening processes.
Figures


References
-
- Lorenz P, Schleper C. Metagenome—a challenging source of enzyme discovery. J Mol Catal B Enzym. 2002;19–20:13–19. doi: 10.1016/S1381-1177(02)00147-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

