Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES
- PMID: 26783202
- PMCID: PMC4797288
- DOI: 10.1093/nar/gkw016
Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES
Abstract
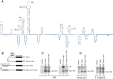
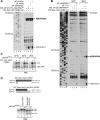
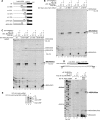
Halastavi árva virus (HalV) has a positive-sense RNA genome, with an 827 nt-long 5' UTR and an intergenic region separating two open reading frames. Whereas the encoded proteins are most homologous to Dicistrovirus polyproteins, its 5' UTR is distinct. Here, we report that the HalV 5' UTR comprises small stem-loop domains separated by long single-stranded areas and a large A-rich unstructured region surrounding the initiation codon AUG828, and possesses cross-kingdom internal ribosome entry site (IRES) activity. In contrast to most viral IRESs, it does not depend on structural integrity and specific interaction of a structured element with a translational component, and is instead determined by the unstructured region flanking AUG828. eIF2, eIF3, eIF1 and eIF1A promote efficient 48S initiation complex formation at AUG828, which is reduced ∼5-fold on omission of eIF1 and eIF1A. Initiation involves direct attachment of 43S preinitiation complexes within a short window at or immediately downstream of AUG828. 40S and eIF3 are sufficient for initial binding. After attachment, 43S complexes undergo retrograde scanning, strongly dependent on eIF1 and eIF1A. eIF4A/eIF4G stimulated initiation only at low temperatures or on mutants, in which areas surrounding AUG828 had been replaced by heterologous sequences. However, they strongly promoted initiation at AUG872, yielding a proline-rich oligopeptide.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

