Human Engineered Cardiac Tissues Created Using Induced Pluripotent Stem Cells Reveal Functional Characteristics of BRAF-Mediated Hypertrophic Cardiomyopathy
- PMID: 26784941
- PMCID: PMC4718533
- DOI: 10.1371/journal.pone.0146697
Human Engineered Cardiac Tissues Created Using Induced Pluripotent Stem Cells Reveal Functional Characteristics of BRAF-Mediated Hypertrophic Cardiomyopathy
Abstract
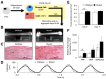
Hypertrophic cardiomyopathy (HCM) is a leading cause of sudden cardiac death that often goes undetected in the general population. HCM is also prevalent in patients with cardio-facio-cutaneous syndrome (CFCS), which is a genetic disorder characterized by aberrant signaling in the RAS/MAPK signaling cascade. Understanding the mechanisms of HCM development in such RASopathies may lead to novel therapeutic strategies, but relevant experimental models of the human condition are lacking. Therefore, the objective of this study was to develop the first 3D human engineered cardiac tissue (hECT) model of HCM. The hECTs were created using human cardiomyocytes obtained by directed differentiation of induced pluripotent stem cells derived from a patient with CFCS due to an activating BRAF mutation. The mutant myocytes were directly conjugated at a 3:1 ratio with a stromal cell population to create a tissue of defined composition. Compared to healthy patient control hECTs, BRAF-hECTs displayed a hypertrophic phenotype by culture day 6, with significantly increased tissue size, twitch force, and atrial natriuretic peptide (ANP) gene expression. Twitch characteristics reflected increased contraction and relaxation rates and shorter twitch duration in BRAF-hECTs, which also had a significantly higher maximum capture rate and lower excitation threshold during electrical pacing, consistent with a more arrhythmogenic substrate. By culture day 11, twitch force was no longer different between BRAF and wild-type hECTs, revealing a temporal aspect of disease modeling with tissue engineering. Principal component analysis identified diastolic force as a key factor that changed from day 6 to day 11, supported by a higher passive stiffness in day 11 BRAF-hECTs. In summary, human engineered cardiac tissues created from BRAF mutant cells recapitulated, for the first time, key aspects of the HCM phenotype, offering a new in vitro model for studying intrinsic mechanisms and screening new therapeutic approaches for this lethal form of heart disease.
Conflict of interest statement
Figures


References
-
- Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 2002;11:2499–506. - PubMed
-
- Schoenfeld JR, Vasser M, Jhurani P, Ng P, Hunter JJ, Ross J, et al. Distinct molecular phenotypes in murine cardiac muscle development, growth, and hypertrophy. Journal of Molecular and Cellular Cardiology 1998;30:2269–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

