Oligoadenylation of 3' decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells
- PMID: 26786835
- PMCID: PMC4748820
- DOI: 10.1261/rna.053942.115
Oligoadenylation of 3' decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells
Abstract
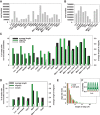
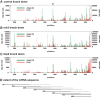
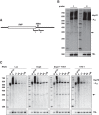
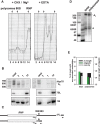
Post-transcriptional 3' end addition of nucleotides is important in a variety of RNA decay pathways. We have examined the 3' end addition of nucleotides during the decay of the Hsp70 mRNA and a corresponding reporter RNA in Drosophila S2 cells by conventional sequencing of cDNAs obtained after mRNA circularization and by deep sequencing of dedicated libraries enriched for 3' decay intermediates along the length of the mRNA. Approximately 5%-10% of 3' decay intermediates carried nonencoded oligo(A) tails with a mean length of 2-3 nucleotides. RNAi experiments showed that the oligoadenylated RNA fragments were intermediates of exosomal decay and the noncanonical poly(A) polymerase Trf4-1 was mainly responsible for A addition. A hot spot of A addition corresponded to an intermediate of 3' decay that accumulated upon inhibition of decapping, and knockdown of Trf4-1 increased the abundance of this intermediate, suggesting that oligoadenylation facilitates 3' decay. Oligoadenylated 3' decay intermediates were found in the cytoplasmic fraction in association with ribosomes, and fluorescence microscopy revealed a cytoplasmic localization of Trf4-1. Thus, oligoadenylation enhances exosomal mRNA degradation in the cytoplasm.
Keywords: exosome; mRNA decay; noncanonical poly(A) polymerase; oligoadenylation.
© 2016 Harnisch et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
