Analysis of spatial-temporal gene expression patterns reveals dynamics and regionalization in developing mouse brain
- PMID: 26786896
- PMCID: PMC4726224
- DOI: 10.1038/srep19274
Analysis of spatial-temporal gene expression patterns reveals dynamics and regionalization in developing mouse brain
Abstract
Allen Brain Atlas (ABA) provides a valuable resource of spatial/temporal gene expressions in mammalian brains. Despite rich information extracted from this database, current analyses suffer from several limitations. First, most studies are either gene-centric or region-centric, thus are inadequate to capture the superposition of multiple spatial-temporal patterns. Second, standard tools of expression analysis such as matrix factorization can capture those patterns but do not explicitly incorporate spatial dependency. To overcome those limitations, we proposed a computational method to detect recurrent patterns in the spatial-temporal gene expression data of developing mouse brains. We demonstrated that regional distinction in brain development could be revealed by localized gene expression patterns. The patterns expressed in the forebrain, medullary and pontomedullary, and basal ganglia are enriched with genes involved in forebrain development, locomotory behavior, and dopamine metabolism respectively. In addition, the timing of global gene expression patterns reflects the general trends of molecular events in mouse brain development. Furthermore, we validated functional implications of the inferred patterns by showing genes sharing similar spatial-temporal expression patterns with Lhx2 exhibited differential expression in the embryonic forebrains of Lhx2 mutant mice. These analysis outcomes confirm the utility of recurrent expression patterns in studying brain development.
Figures
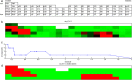

 p-values, black patches indicate low
p-values, black patches indicate low  p-values, and white patches indicate enrichment p-values are not available. The GO categories are divided into four classes: developmental genes, transcription factors, neural information processing and immune response genes. The GO categories in these classes are separated by blue lines.
p-values, and white patches indicate enrichment p-values are not available. The GO categories are divided into four classes: developmental genes, transcription factors, neural information processing and immune response genes. The GO categories in these classes are separated by blue lines.

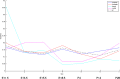
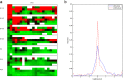
 (forebrain tissue expression ratios) between the Lhx2
(forebrain tissue expression ratios) between the Lhx2
 mutant and the wildtype control among the gene groups possessing similar (blue solid line) and opposite (red dashed line) expression patterns of Lhx2 in the forebrain.
mutant and the wildtype control among the gene groups possessing similar (blue solid line) and opposite (red dashed line) expression patterns of Lhx2 in the forebrain.References
-
- Lein E. S. et al.. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

