4Pipe4--A 454 data analysis pipeline for SNP detection in datasets with no reference sequence or strain information
- PMID: 26787189
- PMCID: PMC4719533
- DOI: 10.1186/s12859-016-0892-1
4Pipe4--A 454 data analysis pipeline for SNP detection in datasets with no reference sequence or strain information
Abstract
Background: Next-generation sequencing datasets are becoming more frequent, and their use in population studies is becoming widespread. For non-model species, without a reference genome, it is possible from a panel of individuals to identify a set of SNPs that can be used for further population genotyping. However the lack of a reference genome to which the sequenced data could be compared makes the finding of SNPs more troublesome. Additionally when the data sources (strains) are not identified (e.g. in datasets of pooled individuals), the problem of finding reliable variation in these datasets can become much more difficult due to the lack of specialized software for this specific task.
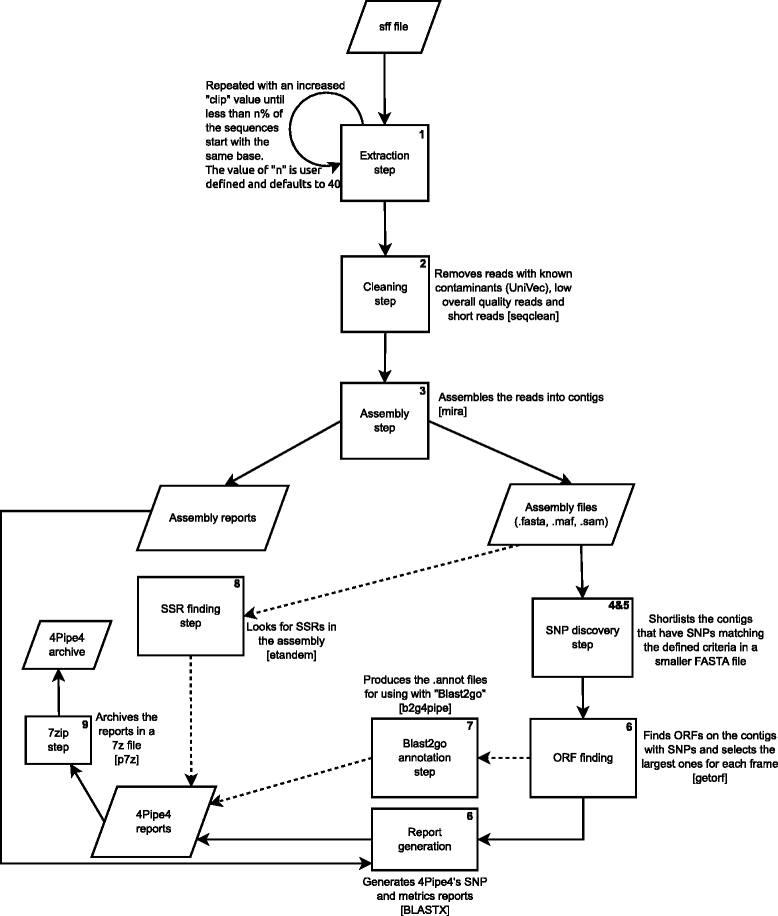
Results: Here we describe 4Pipe4, a 454 data analysis pipeline particularly focused on SNP detection when no reference or strain information is available. It uses a command line interface to automatically call other programs, parse their outputs and summarize the results. The variation detection routine is built-in in the program itself. Despite being optimized for SNP mining in 454 EST data, it is flexible enough to automate the analysis of genomic data or even data from other NGS technologies. 4Pipe4 will output several HTML formatted reports with metrics on many of the most common assembly values, as well as on all the variation found. There is also a module available for finding putative SSRs in the analysed datasets.
Conclusions: This program can be especially useful for researchers that have 454 datasets of a panel of pooled individuals and want to discover and characterize SNPs for subsequent individual genotyping with customized genotyping arrays. In comparison with other SNP detection approaches, 4Pipe4 showed the best validation ratio, retrieving a smaller number of SNPs but with a considerably lower false positive rate than other methods. 4Pipe4's source code is available at https://github.com/StuntsPT/4Pipe4.
Figures
References
-
- Peterlongo P, Schnel N, Pisanti N, Sagot MF, Lacroix V. Identifying SNPs without a Reference Genome by Comparing Raw Reads. In: Chaves E, Lonardi S, editors. String Processing and Information Retrieval. Springer Berlin Heidelberg; 2010. p. 147-58.
-
- Modesto IS, Miguel C, Pina-Martins F, Glushkova M, Veloso M, Paulo OS, et al. Identifying signatures of natural selection in cork oak (Quercus suber L.) genes through SNP analysis. Tree Genet Genomes. 2014;10:1645–60. doi: 10.1007/s11295-014-0786-1. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

