Multigene testing of moderate-risk genes: be mindful of the missense
- PMID: 26787654
- PMCID: PMC4893078
- DOI: 10.1136/jmedgenet-2015-103398
Multigene testing of moderate-risk genes: be mindful of the missense
Abstract
Background: Moderate-risk genes have not been extensively studied, and missense substitutions in them are generally returned to patients as variants of uncertain significance lacking clearly defined risk estimates. The fraction of early-onset breast cancer cases carrying moderate-risk genotypes and quantitative methods for flagging variants for further analysis have not been established.
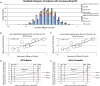
Methods: We evaluated rare missense substitutions identified from a mutation screen of ATM, CHEK2, MRE11A, RAD50, NBN, RAD51, RINT1, XRCC2 and BARD1 in 1297 cases of early-onset breast cancer and 1121 controls via scores from Align-Grantham Variation Grantham Deviation (GVGD), combined annotation dependent depletion (CADD), multivariate analysis of protein polymorphism (MAPP) and PolyPhen-2. We also evaluated subjects by polygenotype from 18 breast cancer risk SNPs. From these analyses, we estimated the fraction of cases and controls that reach a breast cancer OR≥2.5 threshold.
Results: Analysis of mutation screening data from the nine genes revealed that 7.5% of cases and 2.4% of controls were carriers of at least one rare variant with an average OR≥2.5. 2.1% of cases and 1.2% of controls had a polygenotype with an average OR≥2.5.
Conclusions: Among early-onset breast cancer cases, 9.6% had a genotype associated with an increased risk sufficient to affect clinical management recommendations. Over two-thirds of variants conferring this level of risk were rare missense substitutions in moderate-risk genes. Placement in the estimated OR≥2.5 group by at least two of these missense analysis programs should be used to prioritise variants for further study. Panel testing often creates more heat than light; quantitative approaches to variant prioritisation and classification may facilitate more efficient clinical classification of variants.
Keywords: Cancer: breast; Clinical genetics; Genetics; Screening.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
