Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA
- PMID: 26787831
- PMCID: PMC4725013
- DOI: 10.1128/mBio.01981-15
Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA
Abstract
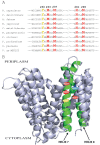
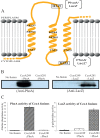
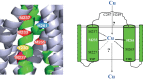
Uptake and trafficking of metals and their delivery to their respective metalloproteins are important processes. Cells need precise control of each step to avoid exposure to excessive metal concentrations and their harmful consequences. Copper (Cu) is a required micronutrient used as a cofactor in proteins. However, in large amounts, it can induce oxidative damage; hence, Cu homeostasis is indispensable for cell survival. Biogenesis of respiratory heme-Cu oxygen (HCO) reductases includes insertion of Cu into their catalytic subunits to form heme-Cu binuclear centers. Previously, we had shown that CcoA is a major facilitator superfamily (MFS)-type bacterial Cu importer required for biogenesis of cbb3-type cytochrome c oxidase (cbb3-Cox). Here, using Rhodobacter capsulatus, we focused on the import and delivery of Cu to cbb3-Cox. By comparing the CcoA amino acid sequence with its homologues from other bacterial species, we located several well-conserved Met, His, and Tyr residues that might be important for Cu transport. We determined the topology of the transmembrane helices that carry these residues to establish that they are membrane embedded, and substituted for them amino acids that do not ligand metal atoms. Characterization of these mutants for their uptake of radioactive (64)Cu and cbb3-Cox activities demonstrated that Met233 and His261 of CcoA are essential and Met237 and Met265 are important, whereas Tyr230 has no role for Cu uptake or cbb3-Cox biogenesis. These findings show for the first time that CcoA-mediated Cu import relies on conserved Met and His residues that could act as metal ligands at the membrane-embedded Cu binding domain of this transporter.
Importance: Cu is a micronutrient that is both essential and toxic; hence, its cellular homeostasis is crucial. Respiratory cbb3-type cytochrome c oxidases (cbb3-Cox) are Cu-containing energy-transducing enzymes that are important for many microaerophilic processes, including photosynthesis, respiration, and bacterial pathogenesis. How Cu is incorporated into cbb3-Cox enzymes is not well known. So far, CcoA is the only known major facilitator superfamily (MFS)-type transporter required for Cu import into the bacterial cytoplasm and for cbb3-Cox biogenesis. This study shows that the membrane-embedded, universally conserved Met and His residues of CcoA are essential for its Cu import function and also for its role in cbb3-Cox biogenesis, shedding light on the mechanism of function of this bacterial prototypical Cu importer.
Copyright © 2016 Khalfaoui-Hassani et al.
Figures

References
-
- Yoshikawa S, Shimada A, Shinzawa-Itoh K. 2015. Respiratory conservation of energy with dioxygen: cytochrome c oxidase. Met Ions Life Sci 15:89–130. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
