Mobile small RNAs regulate genome-wide DNA methylation
- PMID: 26787884
- PMCID: PMC4760824
- DOI: 10.1073/pnas.1515072113
Mobile small RNAs regulate genome-wide DNA methylation
Abstract
RNA silencing at the transcriptional and posttranscriptional levels regulates endogenous gene expression, controls invading transposable elements (TEs), and protects the cell against viruses. Key components of the mechanism are small RNAs (sRNAs) of 21-24 nt that guide the silencing machinery to their nucleic acid targets in a nucleotide sequence-specific manner. Transcriptional gene silencing is associated with 24-nt sRNAs and RNA-directed DNA methylation (RdDM) at cytosine residues in three DNA sequence contexts (CG, CHG, and CHH). We previously demonstrated that 24-nt sRNAs are mobile from shoot to root in Arabidopsis thaliana and confirmed that they mediate DNA methylation at three sites in recipient cells. In this study, we extend this finding by demonstrating that RdDM of thousands of loci in root tissues is dependent upon mobile sRNAs from the shoot and that mobile sRNA-dependent DNA methylation occurs predominantly in non-CG contexts. Mobile sRNA-dependent non-CG methylation is largely dependent on the DOMAINS REARRANGED METHYLTRANSFERASES 1/2 (DRM1/DRM2) RdDM pathway but is independent of the CHROMOMETHYLASE (CMT)2/3 DNA methyltransferases. Specific superfamilies of TEs, including those typically found in gene-rich euchromatic regions, lose DNA methylation in a mutant lacking 22- to 24-nt sRNAs (dicer-like 2, 3, 4 triple mutant). Transcriptome analyses identified a small number of genes whose expression in roots is associated with mobile sRNAs and connected to DNA methylation directly or indirectly. Finally, we demonstrate that sRNAs from shoots of one accession move across a graft union and target DNA methylation de novo at normally unmethylated sites in the genomes of root cells from a different accession.
Keywords: RNA-directed DNA methylation; plant grafting; small RNA; transcriptional gene silencing; transposable element.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

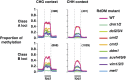
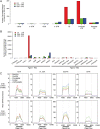
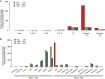

Comment in
-
Systemic silencing: Mobile sRNA stabilizes genomes.Nat Plants. 2016 Mar 2;2:16020. doi: 10.1038/nplants.2016.20. Nat Plants. 2016. PMID: 27249354 No abstract available.
References
-
- Martínez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: A diversity of pathways. Biochim Biophys Acta. 2013;1829(12):1300–1308. - PubMed
-
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. - PubMed
-
- Qi Y, et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443(7114):1008–1012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

