Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers
- PMID: 26787895
- PMCID: PMC4747776
- DOI: 10.1073/pnas.1518752113
Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers
Abstract
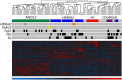
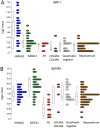
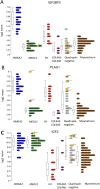
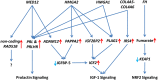
Uterine leiomyomas are common benign smooth muscle tumors that impose a major burden on women's health. Recent sequencing studies have revealed recurrent and mutually exclusive mutations in leiomyomas, suggesting the involvement of molecularly distinct pathways. In this study, we explored transcriptional differences among leiomyomas harboring different genetic drivers, including high mobility group AT-hook 2 (HMGA2) rearrangements, mediator complex subunit 12 (MED12) mutations, biallelic inactivation of fumarate hydratase (FH), and collagen, type IV, alpha 5 and collagen, type IV, alpha 6 (COL4A5-COL4A6) deletions. We also explored the transcriptional consequences of 7q22, 22q, and 1p deletions, aiming to identify possible target genes. We investigated 94 leiomyomas and 60 corresponding myometrial tissues using exon arrays, whole genome sequencing, and SNP arrays. This integrative approach revealed subtype-specific expression changes in key driver pathways, including Wnt/β-catenin, Prolactin, and insulin-like growth factor (IGF)1 signaling. Leiomyomas with HMGA2 aberrations displayed highly significant up-regulation of the proto-oncogene pleomorphic adenoma gene 1 (PLAG1), suggesting that HMGA2 promotes tumorigenesis through PLAG1 activation. This was supported by the identification of genetic PLAG1 alterations resulting in expression signatures as seen in leiomyomas with HMGA2 aberrations. RAD51 paralog B (RAD51B), the preferential translocation partner of HMGA2, was up-regulated in MED12 mutant lesions, suggesting a role for this gene in the genesis of leiomyomas. FH-deficient leiomyomas were uniquely characterized by activation of nuclear factor erythroid 2-related factor 2 (NRF2) target genes, supporting the hypothesis that accumulation of fumarate leads to activation of the oncogenic transcription factor NRF2. This study emphasizes the need for molecular stratification in leiomyoma research and possibly in clinical practice as well. Further research is needed to determine whether the candidate biomarkers presented herein can provide guidance for managing the millions of patients affected by these lesions.
Keywords: HMGA2; MED12; transcriptional profiling; uterine leiomyoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Rice KE, Secrist JR, Woodrow EL, Hallock LM, Neal JL. Etiology, diagnosis, and management of uterine leiomyomas. J Midwifery Womens Health. 2012;57(3):241–247. - PubMed
-
- Zhao D, Rogers PAW. Is fibroid heterogeneity a significant issue for clinicians and researchers? Reprod Biomed Online. 2013;27(1):64–74. - PubMed
-
- Mäkinen N, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334(6053):252–255. - PubMed
-
- Mehine M, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369(1):43–53. - PubMed
-
- Mehine M, Mäkinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: Insights from high-throughput sequencing. Fertil Steril. 2014;102(3):621–629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

