Structural homology guided alignment of cysteine rich proteins
- PMID: 26788439
- PMCID: PMC4709342
- DOI: 10.1186/s40064-015-1609-z
Structural homology guided alignment of cysteine rich proteins
Abstract
Background: Cysteine rich protein families are notoriously difficult to align due to low sequence identity and frequent insertions and deletions.
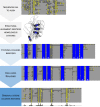
Results: Here we present an alignment method that ensures homologous cysteines align by assigning a unique 10 amino acid barcode to those identified as structurally homologous by the DALI webserver. The free inter-cysteine regions of the barcoded sequences can then be aligned using any standard algorithm. Finally the barcodes are replaced with the original columns to yield an alignment which requires the minimum of manual refinement.
Conclusions: Using structural homology information to constrain sequence alignments allows the alignment of highly divergent, repetitive sequences that are poorly dealt with by existing algorithms. Tools are provided to perform this method online using the CysBar web-tool (http://CysBar.science.latrobe.edu.au) and offline (python script available from http://github.com/ts404/CysBar).
Keywords: Alignment; Barcode; Cysteine-rich proteins; Defensin.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

