Cooperative TRAIL production mediates IFNα/Smac mimetic-induced cell death in TNFα-resistant solid cancer cells
- PMID: 26788912
- PMCID: PMC4826164
- DOI: 10.18632/oncotarget.6915
Cooperative TRAIL production mediates IFNα/Smac mimetic-induced cell death in TNFα-resistant solid cancer cells
Abstract
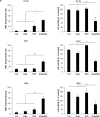
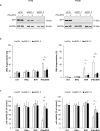
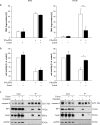
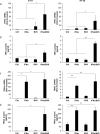
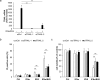
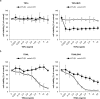
Smac mimetics antagonize IAP proteins, which are highly expressed in several cancers. Recent reports indicate that Smac mimetics trigger a broad cytokine response and synergize with immune modulators to induce cell death. Here, we identify a differential requirement of TRAIL or TNFα as mediators of IFNα/Smac mimetic-induced cell death depending on the cellular context. Subtoxic concentrations of Smac mimetics cooperate with IFNα to induce cell death in various solid tumor cell lines in a highly synergistic manner as determined by combination index. Mechanistic studies show that IFNα/BV6 cotreatment promotes the formation of a caspase-8-activating complex together with the adaptor protein FADD and RIP1. Assembly of this RIP1/FADD/caspase-8 complex represents a critical event, since RIP1 silencing inhibits IFNα/BV6-induced cell death. Strikingly, pharmacological inhibition of paracrine/autocrine TNFα signaling by the TNFα scavenger Enbrel rescues HT-29 colon carcinoma cells, but not A172 glioblastoma cells from IFNα/BV6-induced cell death. By comparison, A172 cells are significantly protected against IFNα/BV6 treatment by blockage of TRAIL signaling through genetic silencing of TRAIL or its cognate receptor TRAIL receptor 2 (DR5). Despite this differential requirement of TNFα and TRAIL signaling, mRNA and protein expression is increased by IFNα/BV6 cotreatment in both cell lines. Interestingly, A172 cells turn out to be resistant to exogenously added recombinant TNFα even in the presence of BV6, whereas they display a high sensitivity towards TRAIL/BV6. In contrast, BV6 efficiently sensitizes HT-29 cells to TNFα while TRAIL only had limited efficacy. This demonstrates that a differential sensitivity towards TRAIL or TNFα determines the dependency on either death receptor ligand for IFNα/Smac mimetic-induced cell death. Thus, by concomitant stimulation of both death receptor systems IFNα/Smac mimetic combination treatment is an effective strategy to induce cell death in TNFα- or TRAIL-responsive cancers.
Keywords: Smac; TRAIL; apoptosis; cell death; interferon.
Conflict of interest statement
None to declare.
Figures





Similar articles
-
Smac mimetic sensitizes renal cell carcinoma cells to interferon-α-induced apoptosis.Cancer Lett. 2016 May 28;375(1):1-8. doi: 10.1016/j.canlet.2016.02.019. Epub 2016 Feb 18. Cancer Lett. 2016. PMID: 26912071
-
Synergistic interaction of Smac mimetic and IFNα to trigger apoptosis in acute myeloid leukemia cells.Cancer Lett. 2014 Dec 28;355(2):224-31. doi: 10.1016/j.canlet.2014.08.040. Epub 2014 Aug 29. Cancer Lett. 2014. PMID: 25179908
-
Smac mimetic triggers necroptosis in pancreatic carcinoma cells when caspase activation is blocked.Cancer Lett. 2016 Sep 28;380(1):31-8. doi: 10.1016/j.canlet.2016.05.036. Epub 2016 Jun 3. Cancer Lett. 2016. PMID: 27267809
-
Therapeutic Small Molecules Target Inhibitor of Apoptosis Proteins in Cancers with Deregulation of Extrinsic and Intrinsic Cell Death Pathways.Clin Cancer Res. 2017 Mar 15;23(6):1379-1387. doi: 10.1158/1078-0432.CCR-16-2172. Epub 2016 Dec 30. Clin Cancer Res. 2017. PMID: 28039268 Free PMC article. Review.
-
Regulation of Cancer Metastasis by TRAIL/Death Receptor Signaling.Biomolecules. 2021 Mar 26;11(4):499. doi: 10.3390/biom11040499. Biomolecules. 2021. PMID: 33810241 Free PMC article. Review.
Cited by
-
A Bak-dependent mitochondrial amplification step contributes to Smac mimetic/glucocorticoid-induced necroptosis.Cell Death Differ. 2017 Jan;24(1):83-97. doi: 10.1038/cdd.2016.102. Epub 2016 Nov 11. Cell Death Differ. 2017. PMID: 27834956 Free PMC article.
-
Oncolytic virus synergizes with Smac mimetic compounds to induce rhabdomyosarcoma cell death in a syngeneic murine model.Oncotarget. 2017 Jan 10;8(2):3495-3508. doi: 10.18632/oncotarget.13849. Oncotarget. 2017. PMID: 27966453 Free PMC article.
-
Cell death-based treatment of glioblastoma.Cell Death Dis. 2018 Jan 25;9(2):121. doi: 10.1038/s41419-017-0021-8. Cell Death Dis. 2018. PMID: 29371590 Free PMC article. Review.
-
Targeting triple-negative breast cancers with the Smac-mimetic birinapant.Cell Death Differ. 2020 Oct;27(10):2768-2780. doi: 10.1038/s41418-020-0541-0. Epub 2020 Apr 27. Cell Death Differ. 2020. PMID: 32341449 Free PMC article.
-
The Smac mimetic BV6 cooperates with STING to induce necroptosis in apoptosis-resistant pancreatic carcinoma cells.Cell Death Dis. 2021 Aug 30;12(9):816. doi: 10.1038/s41419-021-04014-x. Cell Death Dis. 2021. PMID: 34462421 Free PMC article.
References
-
- Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. - PubMed
-
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. - PubMed
-
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. - PubMed
-
- Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

