Universal Solid-Phase Reversible Sample-Prep for Concurrent Proteome and N-Glycome Characterization
- PMID: 26791391
- PMCID: PMC5440117
- DOI: 10.1021/acs.jproteome.5b00865
Universal Solid-Phase Reversible Sample-Prep for Concurrent Proteome and N-Glycome Characterization
Abstract
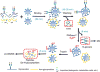
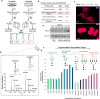
We describe a novel solid-phase reversible sample-prep (SRS) platform that enables rapid sample preparation for concurrent proteome and N-glycome characterization for nearly all protein samples. SRS utilizes a uniquely functionalized, silica-based bead that has strong affinity toward proteins with minimal to no affinity for peptides and other small molecules. By leveraging this inherent size difference between proteins and peptides, SRS permits high-capacity binding of proteins, rapid removal of small molecules (detergents, metabolites, salts, peptides, etc.), extensive manipulation including enzymatic and chemical treatments on bead-bound proteins, and easy recovery of N-glycans and peptides. SRS was evaluated in a wide range of samples including glycoproteins, cell lysate, murine tissues, and human urine. SRS was also coupled to a quantitative strategy to investigate the differences between DU145 prostate cancer cells and its DIAPH3-silenced counterpart. Previous studies suggested that DIAPH3 silencing in DU145 induced transition to an amoeboid phenotype that correlated with tumor progression and metastasis. In this pilot study we identified distinct proteomic and N-glycomic alterations between them. A metastasis-associated tyrosine kinase receptor ephrin-type-A receptor (EPHA2) was highly up-regulated in DIAPH3-silenced cells, indicating a possible connection between EPHA2 and DIAPH3. Moreover, distinct alterations in the N-glycome were identified, suggesting cross-links between DIAPH3 and glycosyltransferase networks.
Keywords: N-glycome; glycoprotein; membrane protein; plasma; proteome; reversible solid phase; sample-prep; urine.
Conflict of interest statement
Boston Children’s Hospital has filed a patent application covering the invention described in this report.
Figures

References
-
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. - PubMed
-
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. - PubMed
-
- Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

