Detection and replication of QTL underlying resistance to gastrointestinal nematodes in adult sheep using the ovine 50K SNP array
- PMID: 26791855
- PMCID: PMC4719203
- DOI: 10.1186/s12711-016-0182-4
Detection and replication of QTL underlying resistance to gastrointestinal nematodes in adult sheep using the ovine 50K SNP array
Abstract
Background: Persistence of gastrointestinal nematode (GIN) infection and the related control methods have major impacts on the sheep industry worldwide. Based on the information generated with the Illumina OvineSNP50 BeadChip (50 K chip), this study aims at confirming quantitative trait loci (QTL) that were previously identified by microsatellite-based genome scans and identifying new QTL and allelic variants that are associated with indicator traits of parasite resistance in adult sheep. We used a commercial half-sib population of 518 Spanish Churra ewes with available data for fecal egg counts (FEC) and serum levels of immunoglobulin A (IgA) to perform different genome scan QTL mapping analyses based on classical linkage analysis (LA), a combined linkage disequilibrium and linkage analysis (LDLA) and a genome-wide association study (GWAS).
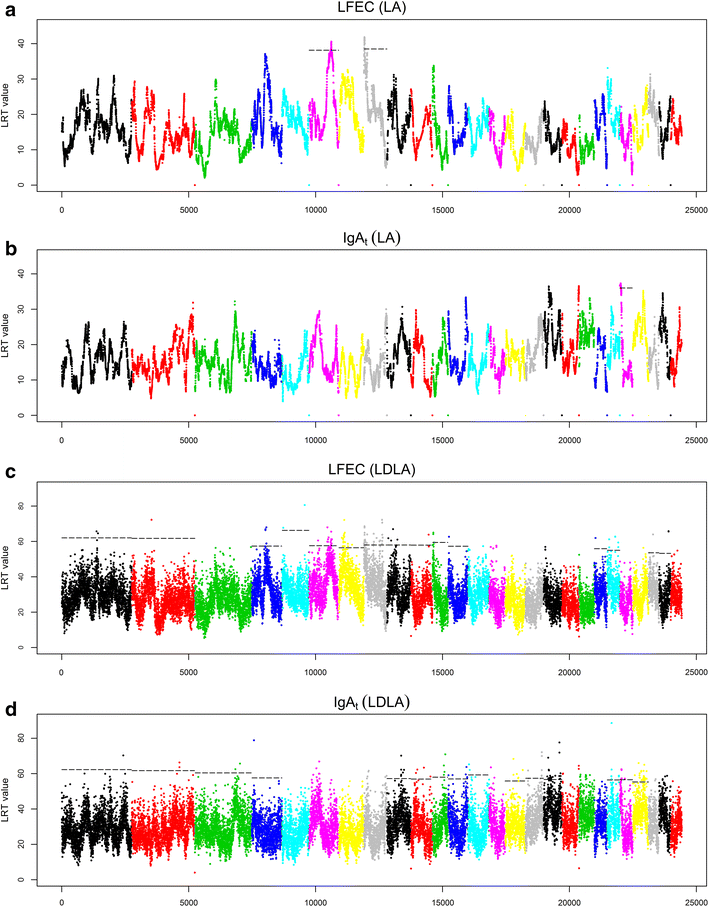
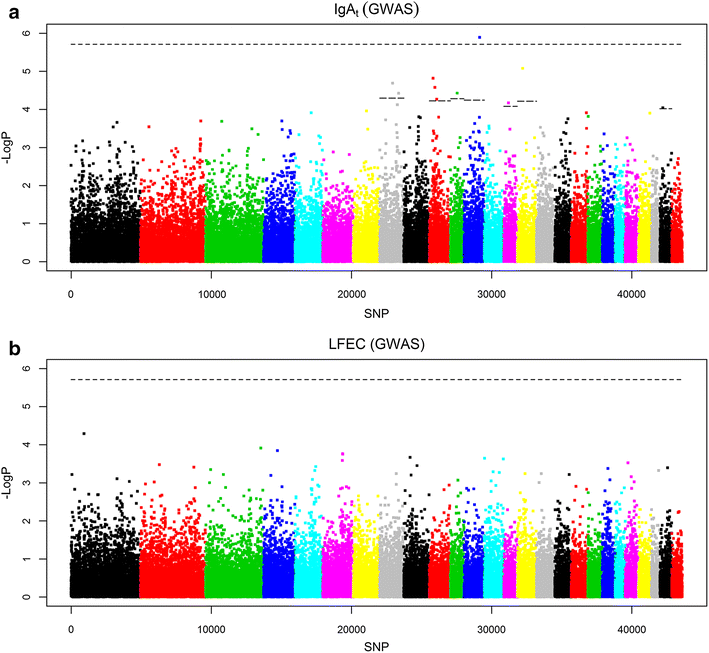
Results: For the FEC and IgA traits, we detected a total of three 5 % chromosome-wise significant QTL by LA and 63 significant regions by LDLA, of which 13 reached the 5 % genome-wise significance level. The GWAS also revealed 10 significant SNPs associated with IgAt, although no significant associations were found for LFEC. Some of the significant QTL for LFEC that were detected by LA and LDLA on OAR6 overlapped with a highly significant QTL that was previously detected in a different half-sib population of Churra sheep. In addition, several new QTL and SNP associations were identified, some of which show correspondence with effects that were reported for different populations of young sheep. Other significant associations that did not coincide with previously reported associations could be related to the specific immune response of adult animals.
Discussion: Our results replicate a FEC-related QTL located on OAR6 that was previously reported in Churra sheep and provide support for future research on the identification of the allelic variant that underlies this QTL. The small proportion of genetic variance explained by the detected QTL and the large number of functional candidate genes identified here are consistent with the hypothesis that GIN resistance/susceptibility is a complex trait that is not determined by individual genes acting alone but rather by complex multi-gene interactions. Future studies that combine genomic variation analysis and functional genomic information may help elucidate the biology of GIN disease resistance in sheep.
Figures
Similar articles
-
Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50K SNP chip and whole-genome sequencing.J Dairy Sci. 2018 Oct;101(10):9072-9088. doi: 10.3168/jds.2018-14736. Epub 2018 Aug 9. J Dairy Sci. 2018. PMID: 30100503
-
Discovery of quantitative trait loci for resistance to parasitic nematode infection in sheep: I. Analysis of outcross pedigrees.BMC Genomics. 2006 Jul 18;7:178. doi: 10.1186/1471-2164-7-178. BMC Genomics. 2006. PMID: 16846521 Free PMC article.
-
Identification of quantitative trait loci underlying milk traits in Spanish dairy sheep using linkage plus combined linkage disequilibrium and linkage analysis approaches.J Dairy Sci. 2013 Sep;96(9):6059-69. doi: 10.3168/jds.2013-6824. Epub 2013 Jun 28. J Dairy Sci. 2013. PMID: 23810588
-
Genomic Regions Associated with Sheep Resistance to Gastrointestinal Nematodes.Trends Parasitol. 2016 Jun;32(6):470-480. doi: 10.1016/j.pt.2016.03.007. Epub 2016 May 13. Trends Parasitol. 2016. PMID: 27183838 Review.
-
Quantitative trait loci for internal nematode resistance in sheep: a review.Genet Sel Evol. 2005;37 Suppl 1(Suppl 1):S83-96. doi: 10.1186/1297-9686-37-S1-S83. Genet Sel Evol. 2005. PMID: 15601597 Free PMC article. Review.
Cited by
-
Association analysis and functional annotation of imputed sequence data within genomic regions influencing resistance to gastro-intestinal parasites detected by an LDLA approach in a nucleus flock of Sarda dairy sheep.Genet Sel Evol. 2022 Jan 3;54(1):2. doi: 10.1186/s12711-021-00690-7. Genet Sel Evol. 2022. PMID: 34979909 Free PMC article.
-
Unveiling genomic regions that underlie differences between Afec-Assaf sheep and its parental Awassi breed.Genet Sel Evol. 2017 Feb 10;49(1):19. doi: 10.1186/s12711-017-0296-3. Genet Sel Evol. 2017. PMID: 28187715 Free PMC article.
-
Genomic scans for selective sweeps through haplotype homozygosity and allelic fixation in 14 indigenous sheep breeds from Middle East and South Asia.Sci Rep. 2021 Feb 2;11(1):2834. doi: 10.1038/s41598-021-82625-2. Sci Rep. 2021. PMID: 33531649 Free PMC article.
-
Genome-wide association study and genomic heritabilities for blood protein levels in Lori-Bakhtiari sheep.Sci Rep. 2021 Dec 9;11(1):23771. doi: 10.1038/s41598-021-03290-z. Sci Rep. 2021. PMID: 34887490 Free PMC article.
-
Identifying coevolving loci using interspecific genetic correlations.Ecol Evol. 2017 Jul 28;7(17):6894-6903. doi: 10.1002/ece3.3107. eCollection 2017 Sep. Ecol Evol. 2017. PMID: 28904769 Free PMC article.
References
-
- Raadsma HW, Gray GD, Woolaston RR. Genetics of disease resistance and vaccine response. In: Piper L, Ruvinsky A, editors. The Genetics of Sheep. University Press: Cambridge; 1997. p. 199–224.
-
- Morris CA, Wheeler M, Watson TG, Hosking BC, Leathwick DM. Direct and correlated responses to selection for high or low faecal nematode egg count in Perendale sheep. NZJ Agric Res. 2005;48:1–10. doi: 10.1080/00288233.2005.9513625. - DOI
-
- Karlsson LJE, Greeff JC. Selection response in fecal worm egg counts in the Rylington Merino parasite resistant flock. Aust J Exp Agric. 2006;46:809–811. doi: 10.1071/EA05367. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

