Forging T-Lymphocyte Identity: Intersecting Networks of Transcriptional Control
- PMID: 26791859
- PMCID: PMC4747653
- DOI: 10.1016/bs.ai.2015.09.002
Forging T-Lymphocyte Identity: Intersecting Networks of Transcriptional Control
Abstract
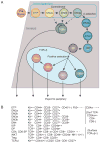
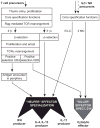
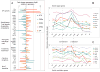
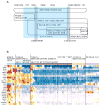
T-lymphocyte development branches off from other lymphoid developmental programs through its requirement for sustained environmental signals through the Notch pathway. In the thymus, Notch signaling induces a succession of T-lineage regulatory factors that collectively create the T-cell identity through distinct steps. This process involves both the staged activation of T-cell identity genes and the staged repression of progenitor-cell-inherited regulatory genes once their roles in self-renewal and population expansion are no longer needed. With the recent characterization of innate lymphoid cells (ILCs) that share transcriptional regulation programs extensively with T-cell subsets, T-cell identity can increasingly be seen as defined in modular terms, as the processes selecting and actuating effector function are potentially detachable from the processes generating and selecting clonally unique T-cell receptor structures. The developmental pathways of different classes of T cells and ILCs are distinguished by the numbers of prerequisites of gene rearrangement, selection, and antigen contact before the cells gain access to nearly common regulatory mechanisms for choosing effector function. Here, the major classes of transcription factors that interact with Notch signals during T-lineage specification are discussed in terms of their roles in these programs, the evidence for their spectra of target genes at different stages, and their cross-regulatory and cooperative actions with each other. Specific topics include Notch modulation of PU.1 and GATA-3, PU.1-Notch competition, the relationship between PU.1 and GATA-3, and the roles of E proteins, Bcl11b, and GATA-3 in guiding acquisition of T-cell identity while avoiding redirection to an ILC fate.
Keywords: E2A; GATA-3; Gene regulation; Notch; PU.1; T-cell development; Transcription factor.
© 2016 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Transcriptional control of T-cell development.Int Immunol. 2011 Nov;23(11):661-8. doi: 10.1093/intimm/dxr078. Epub 2011 Sep 23. Int Immunol. 2011. PMID: 21948191 Review.
-
Repression of Ccr9 transcription in mouse T lymphocyte progenitors by the Notch signaling pathway.J Immunol. 2015 Apr 1;194(7):3191-200. doi: 10.4049/jimmunol.1402443. Epub 2015 Feb 20. J Immunol. 2015. PMID: 25710912 Free PMC article.
-
Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):5800-5807. doi: 10.1073/pnas.1610617114. Proc Natl Acad Sci U S A. 2017. PMID: 28584128 Free PMC article.
-
Transcriptional establishment of cell-type identity: dynamics and causal mechanisms of T-cell lineage commitment.Cold Spring Harb Symp Quant Biol. 2013;78:31-41. doi: 10.1101/sqb.2013.78.020271. Epub 2013 Oct 17. Cold Spring Harb Symp Quant Biol. 2013. PMID: 24135716 Free PMC article. Review.
-
The E-Id Protein Axis Specifies Adaptive Lymphoid Cell Identity and Suppresses Thymic Innate Lymphoid Cell Development.Immunity. 2017 May 16;46(5):818-834.e4. doi: 10.1016/j.immuni.2017.04.022. Immunity. 2017. PMID: 28514688 Free PMC article.
Cited by
-
Fitting structure to function in gene regulatory networks.Hist Philos Life Sci. 2017 Oct 16;39(4):37. doi: 10.1007/s40656-017-0164-z. Hist Philos Life Sci. 2017. PMID: 29038942 Free PMC article.
-
Emerging glycobiology tools: A renaissance in accessibility.Cell Immunol. 2018 Nov;333:2-8. doi: 10.1016/j.cellimm.2018.04.010. Epub 2018 Apr 25. Cell Immunol. 2018. PMID: 29759530 Free PMC article. Review.
-
CD28 Signaling Drives Notch Ligand Expression on CD4 T Cells.Front Immunol. 2020 May 7;11:735. doi: 10.3389/fimmu.2020.00735. eCollection 2020. Front Immunol. 2020. PMID: 32457739 Free PMC article.
-
Native stem cell transcriptional circuits define cardinal features of high-risk leukemia.J Exp Med. 2025 Apr 7;222(4):e20231349. doi: 10.1084/jem.20231349. Epub 2025 Feb 19. J Exp Med. 2025. PMID: 39969525
-
Patterns of Differentially Expressed circRNAs in Human Thymocytes.Noncoding RNA. 2022 Mar 30;8(2):26. doi: 10.3390/ncrna8020026. Noncoding RNA. 2022. PMID: 35447889 Free PMC article.
References
-
- Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A. 2011;108:6211–6216. - PMC - PubMed
-
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. - PubMed
-
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

